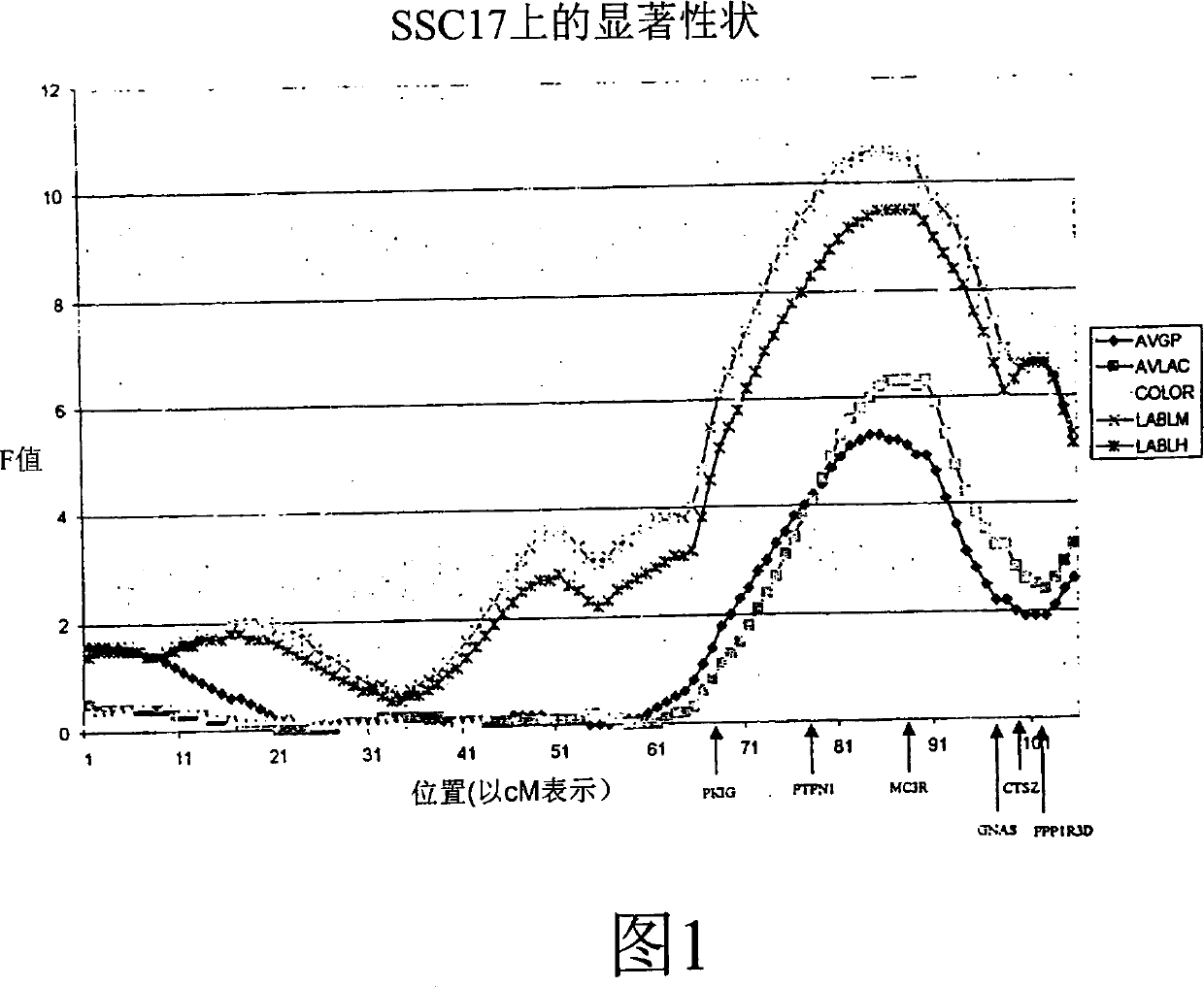
Fine mapping of chromosome 17 quantitative trait loci and use of same for marker assisted selection
A marker-assisted selection, quantitative trait technology, applied in the detection of genetic differences, the field of genetic variation, can solve problems such as thermal stability and ligand affinity reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0186] Cathepsin Z (CTSZ) PCR-RFLP Test
[0187] AlwNI polymorphism
[0188] Primer
[0189] CT04F: 5'GGC ATT TGG GGC ATC TGG G 3' (SEQ ID NO: )
[0190] CT04R: 5' ACT GGG GGA TGT GCT GGT T 3' (SEQ ID NO: )
[0191] PCR conditions:
[0192] 10×Promega buffer
[0193] Combine 10 µL of Mixture 1 and DNA in a reaction tube and cover with mineral oil. The following PCR program was run: 94°C for 3 minutes; 35 cycles of 94°C for 30 seconds, 62°C for 30 seconds and 72°C for 30 seconds; followed by a final extension at 72°C for 5 minutes.
[0194] AlwNI digestion reaction
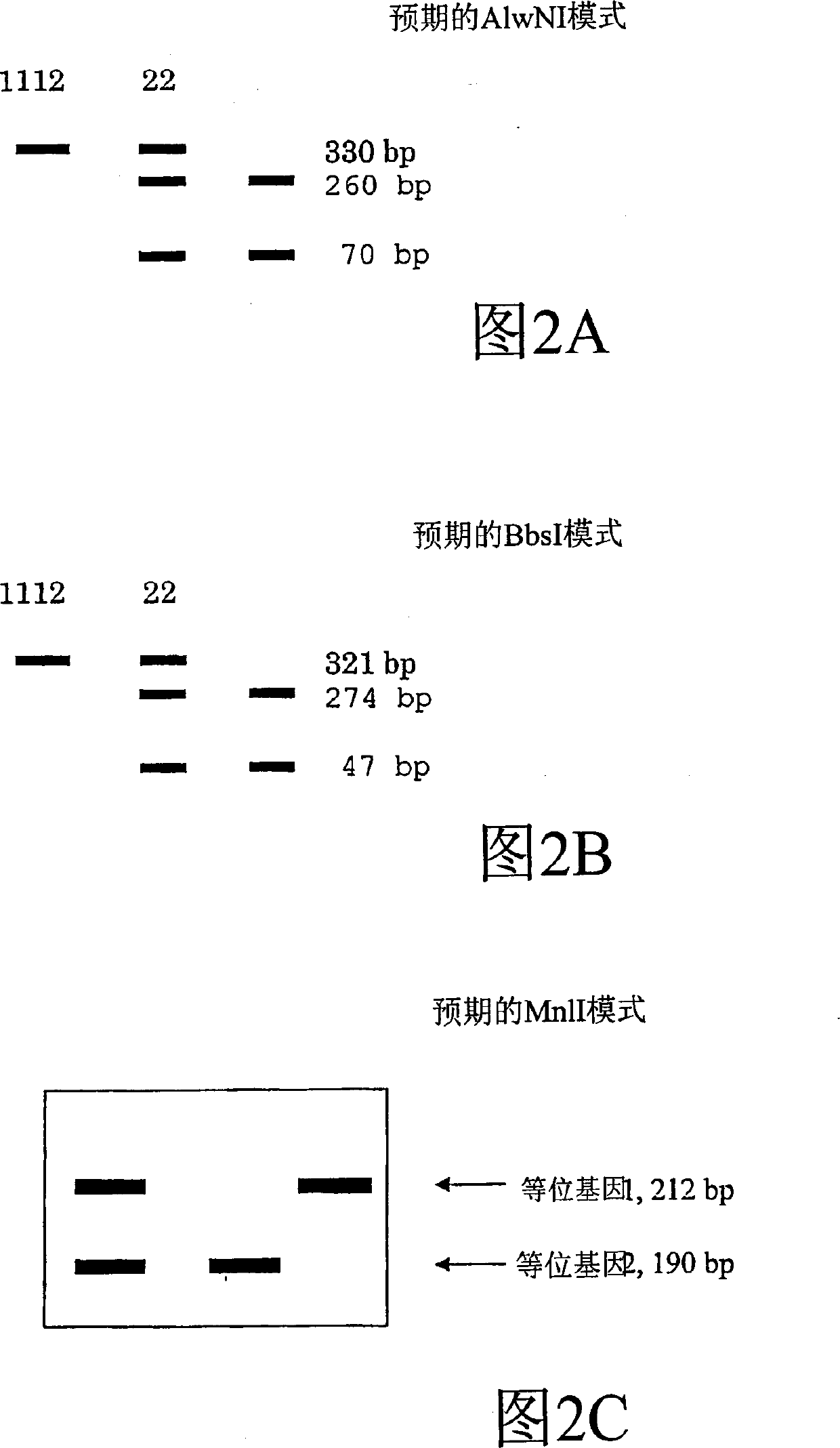
[0195] The buffer, enzyme and water are made into a mixture. Add 5 µL to each reaction tube containing the DNA. Incubate at 37°C for at least 4 hours, preferably digest overnight. 4 μL of loading dye was mixed with 6 μL of digested PCR product and the total volume was loaded on a 3% agarose gel. The desired AlwNI pattern is shown in Figure 2A.
Embodiment 2
[0197] GNAS PCR-RFLP test
[0198] BbsI polymorphism
[0199] A. Primers
[0200] GN03F: 5'AAG CAG GCT GAC TAC GTG 3' (SEQ ID NO: )
[0201] GN03R: 5' TCA CCA CAA GGG CTA CCA 3' (SEQ ID NO: )
[0202] PCR conditions:
[0203] 10×Promega buffer
[0204] Combine 10 µL of Mixture 1 and DNA in a reaction tube and cover with mineral oil. The following PCR program was run: 94°C for 3 minutes; 35 cycles of 94°C for 30 seconds, 60°C for 30 seconds and 72°C for 30 seconds; followed by a final extension at 72°C for 5 minutes.
[0205] BbsI digestion reaction
[0206] The buffer, enzyme and water are made into a mixture. Add 6 µL to each reaction tube containing the DNA. Incubate at 37°C for at least 4 hours, preferably digest overnight. 4 μL of loading dye was mixed with 6 μL of digested PCR product and the total volume was loaded on a 3% agarose gel. The desired BbsI pattern is shown in Figure 2B.
Embodiment 3
[0208] MC3R PCR-RFLP test
[0209] MnlI polymorphism
[0210] Primers:
[0211] Forward: 5'GCC TCC ATC TGC AAC CTC T 3' (SEQ ID NO: )
[0212] Reverse: 5' AGC ATG GCG AAG AAG ATG AC 3' (SEQ ID NO: )
[0213] PCR conditions:
[0214] 10×PCR buffer
[0215]Combine Mixture 1 and DNA in a reaction tube and cover with mineral oil. The following PCR program was run: 3 minutes at 94°C; 36 cycles of 30 seconds at 94°C, 1 minute at 54°C and 30 seconds at 72°C; followed by a final extension at 72°C for 10 minutes.
[0216] 2 [mu]L of the PCR reaction was run on a 1.6% agarose gel to confirm successful amplification and negative controls were cleared. Digestion can be performed using the following procedure:
[0217] PCR product
[0218] Mix PCR product, buffer, enzyme and water. Incubate for at least 4 hours, preferably overnight at 37°C. Digests were mixed with loading dye (2:5) and run on 3% NuSieve agarose gels. The ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com