Fragmentation-based methods and systems for sequence variation detection and discovery
A sequence and mutation technology, applied in the field of fragmentation and system for sequence variation detection and discovery, can solve problems such as complex problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
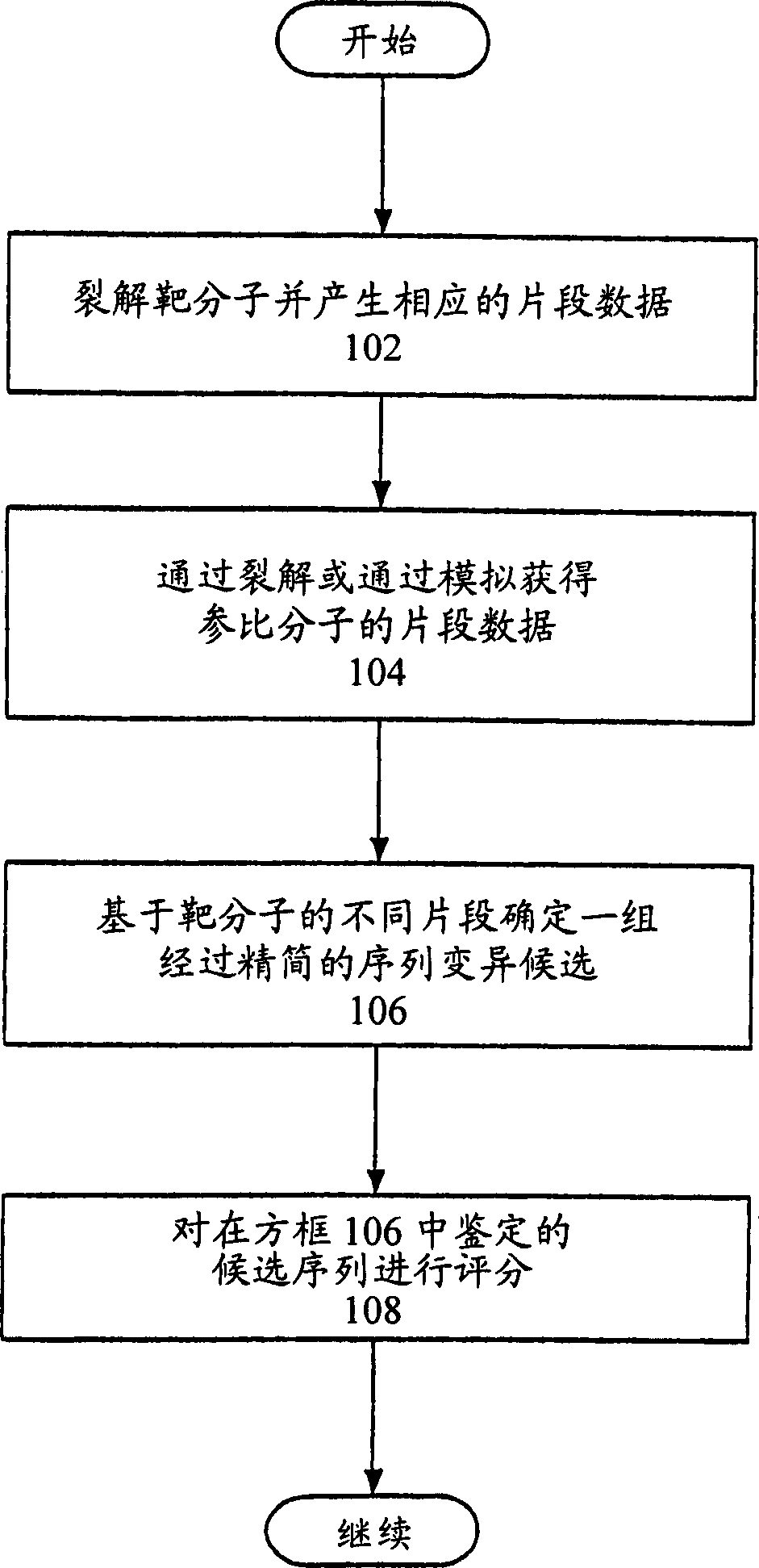
[0388] Base-specific cleavage of RNA
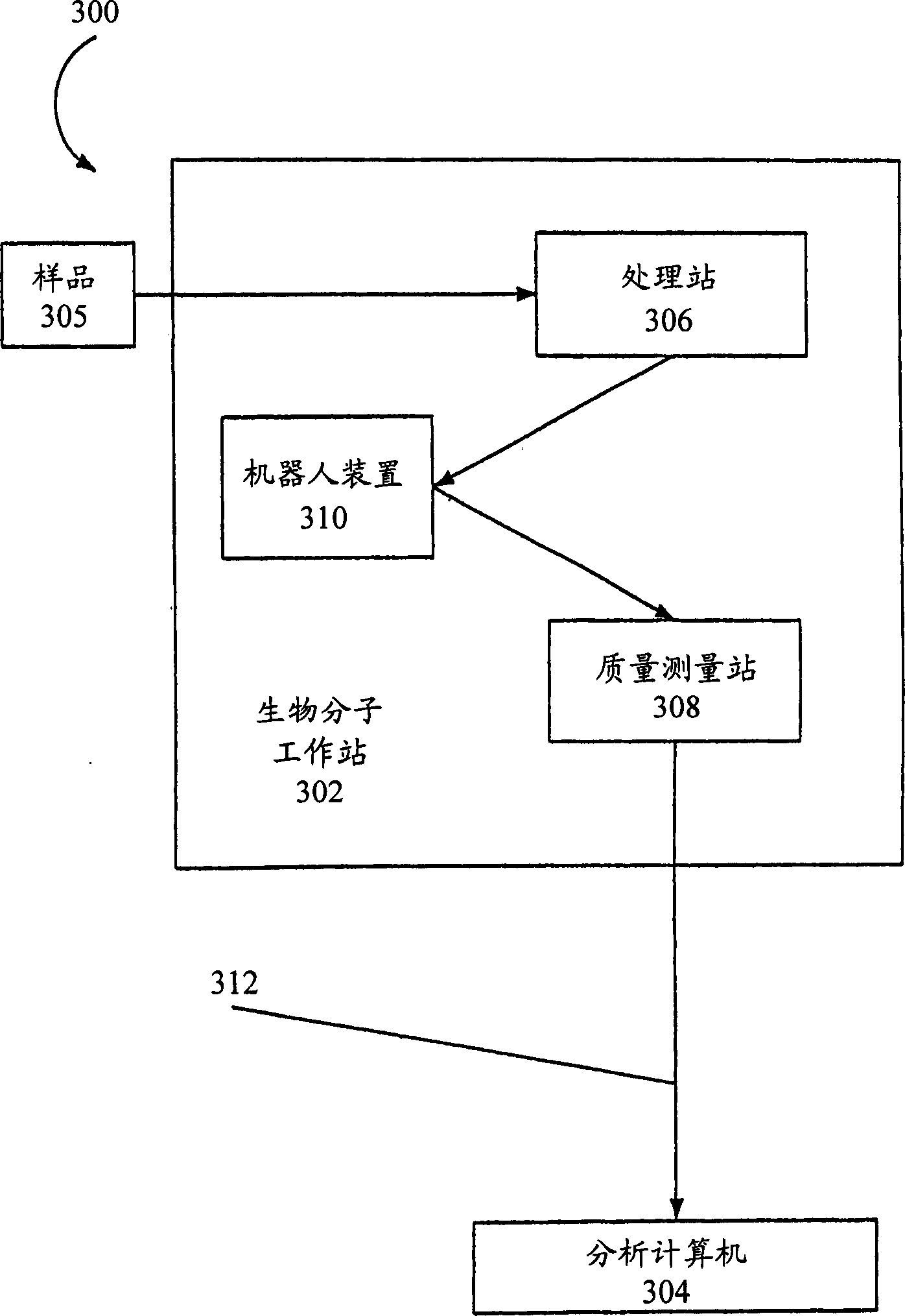
[0389] Presented here is a semi-automated protocol for a one-tube reaction involving RNA transcription and G-specific endonucleolytic cleavage using a typical ribonuclease (RNase T1) to analyze sequence variation in the target nucleic acid of interest . Fragments produced by the ribonuclease cleavage methods provided herein can be analyzed according to the methods provided herein. The RNase T1 reaction is performed to cleave approximately 100% of the G nucleotide site on the target nucleic acid. This cleavage produces a characteristic map of fragment masses that represent sequence variation in the target sequence of interest.
[0390] Materials and methods
[0391] Oligonucleotides were purchased from Metabion (Germany). 5-Methylcytidine lithium 5'-triphosphate (Me-CTP) and 5-methyluridine lithium 5'-triphosphate (Me-UTP) were obtained from Trilink (USA).
[0392] PCR amplification
[0393] A 5 μl PCR reaction included 5 ng of genom...
Embodiment 2
[0430] Base-specific cleavage of DNA
[0431] The following examples describe methods for cleaving a target nucleic acid based on the presence of U residues in the nucleic acid by digestion with uracil DNA glycosylase and the use of NH 3 cleavage of the phosphate backbone. The fragmentation methods provided herein can be used to generate base-specific cleaved fragments of target DNA, which can then be analyzed according to the methods provided herein to identify sequence variations in the target DNA relative to a reference DNA.
[0432] The DNA region of interest was amplified by PCR in the presence of dUTP instead of dTTP. The target region was amplified using a 50 μl PCR reaction consisting of 25 ng of genomic DNA, 1 unit of HotStarTaq DNA polymerase (Qiagen), 0.2 mM each of dATP, dCTP, and dGTP, and 0.6 mM each in 1×HotStarTaq PCR buffer. mM dUTP. PCR primers were used in an asymmetric ratio of 5 pmol biotinylated primer and 15 pmol non-biotinylated primer. The temperat...
Embodiment 3
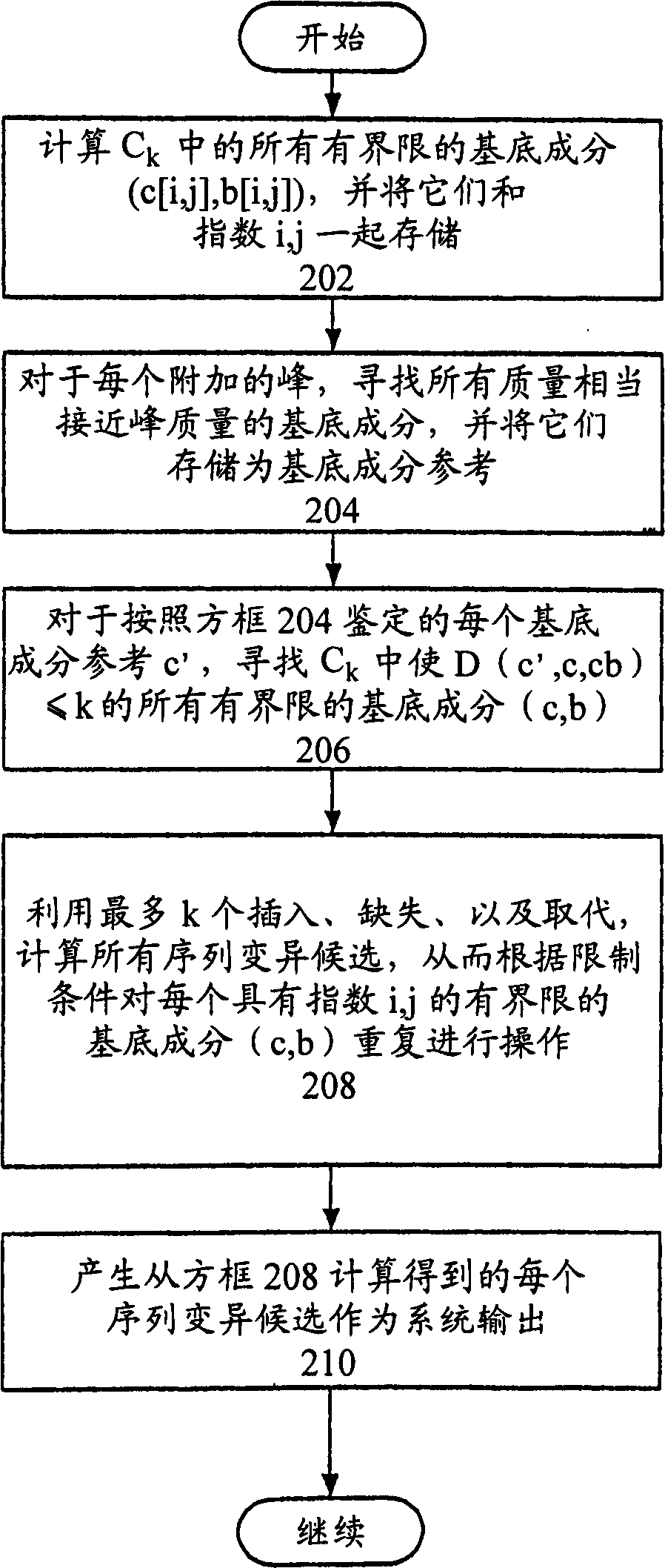
[0451] A. SNP discovery by amplifying base-specific breaks in DNA
[0452] Base-specific cleavage fragments of a target sequence comprising a SNP can be analyzed by the methods provided herein to detect known SNPs or to discover unknown SNPs. High-throughput base-specific fragmentation followed by mass spectrometry can be performed as described by Rodi et al., Bio Techniques, 32:S62-S69 (2002) (incorporated herein by reference), using a system such as that developed by the trademark MassARRAY TM represented system. Mass ARRAY TM Rely on mass spectrometry combined with miniaturized arrays and MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time-of-Flight) mass spectrometry to deliver results rapidly. Fragment signals generated according to the methods provided herein and Rodi et al., Bio Techniques, 32:S62-S69 (2002) can be analyzed according to the methods provided herein.
[0453] In base-specific fragmentation, a single-stranded copy of the target sequence is creat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com