Recombinant serine protease and fungicide containing the same
A technology of serine protease and fungi, which is applied in the direction of nematicides, biocides, fungi, etc., can solve the problems of non-fusion, etc., and achieve the effect of improving virulence and speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Cloning of chitin binding domain
[0039] The amplified template is cDNA reverse-transcribed from the mRNA of the fourth-instar silkworm (provided by the Sericulture College of Southwest Agricultural University) during the molting period (the reverse transcription kit is a product of TaKaRa Company) as a template.
[0040] Primers were designed according to the sequence of silkworm chitinase (GenBank accession number: AB052914):
[0041] P1: 5'-ACT AGT CAC AAC CAC CAC CAC CGT G-3'
[0042] P2: 5'-GCG GCC GCC CGG GTT ACG AAC ATF CCG GTC TGT-3'
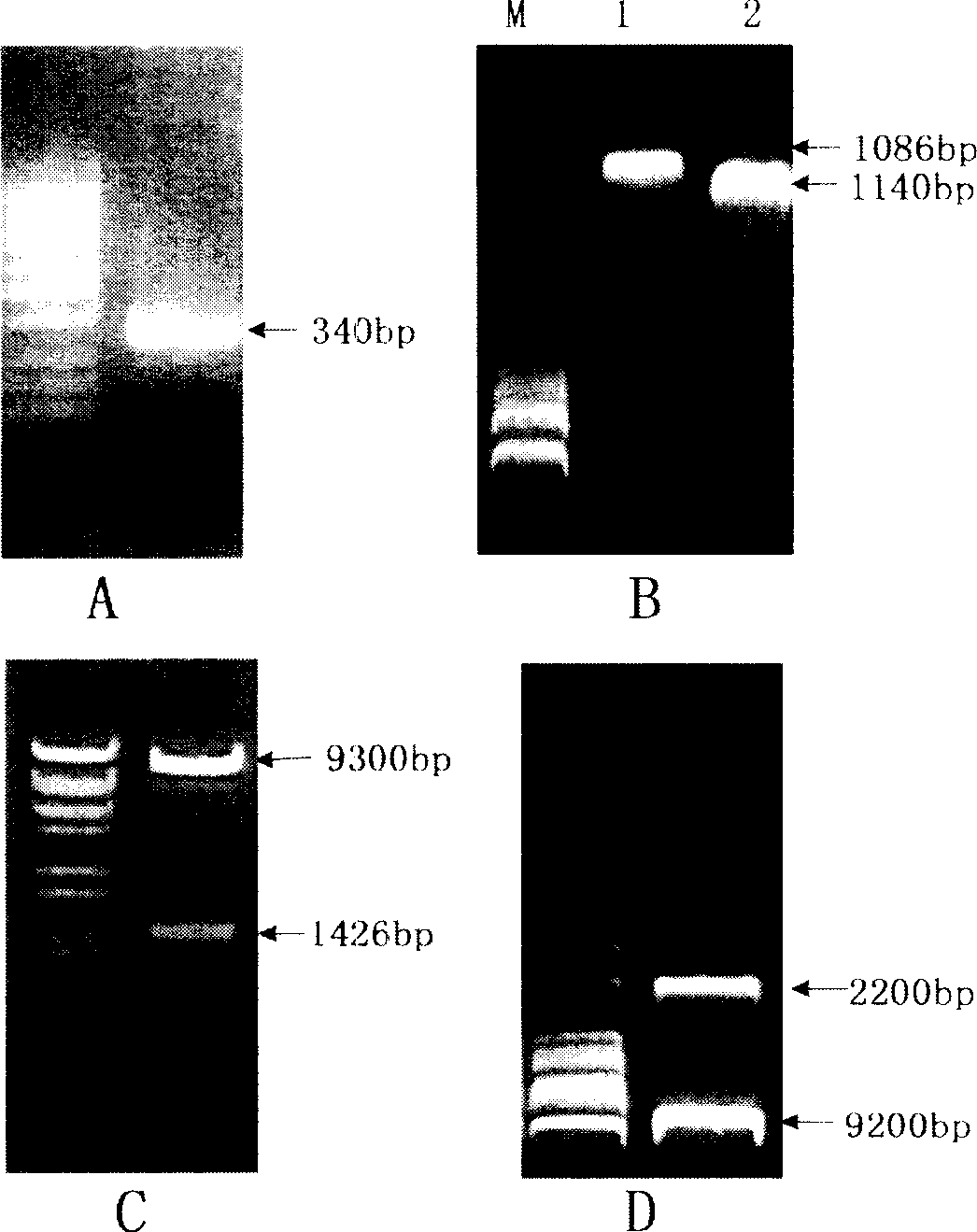
[0043] High-fidelity pfu DNA polymerase (product of Dingguo Company) was used for PCR amplification, and the PCR amplification conditions were as follows: 94°C for 5min; 94°C for 30sec, 58°C for 30sec, 72°C for 1min, 30 cycles; adding 0.7 units of Taq DNA polymerase was extended for another 20 min. A binding domain gene (sequence shown in SEQ ID NO.4) of about 340 bp was obtained by PCR amplification, and corresponding enzy...
Embodiment 2
[0065] 1. Construction of CDEP-1 yeast expression vector
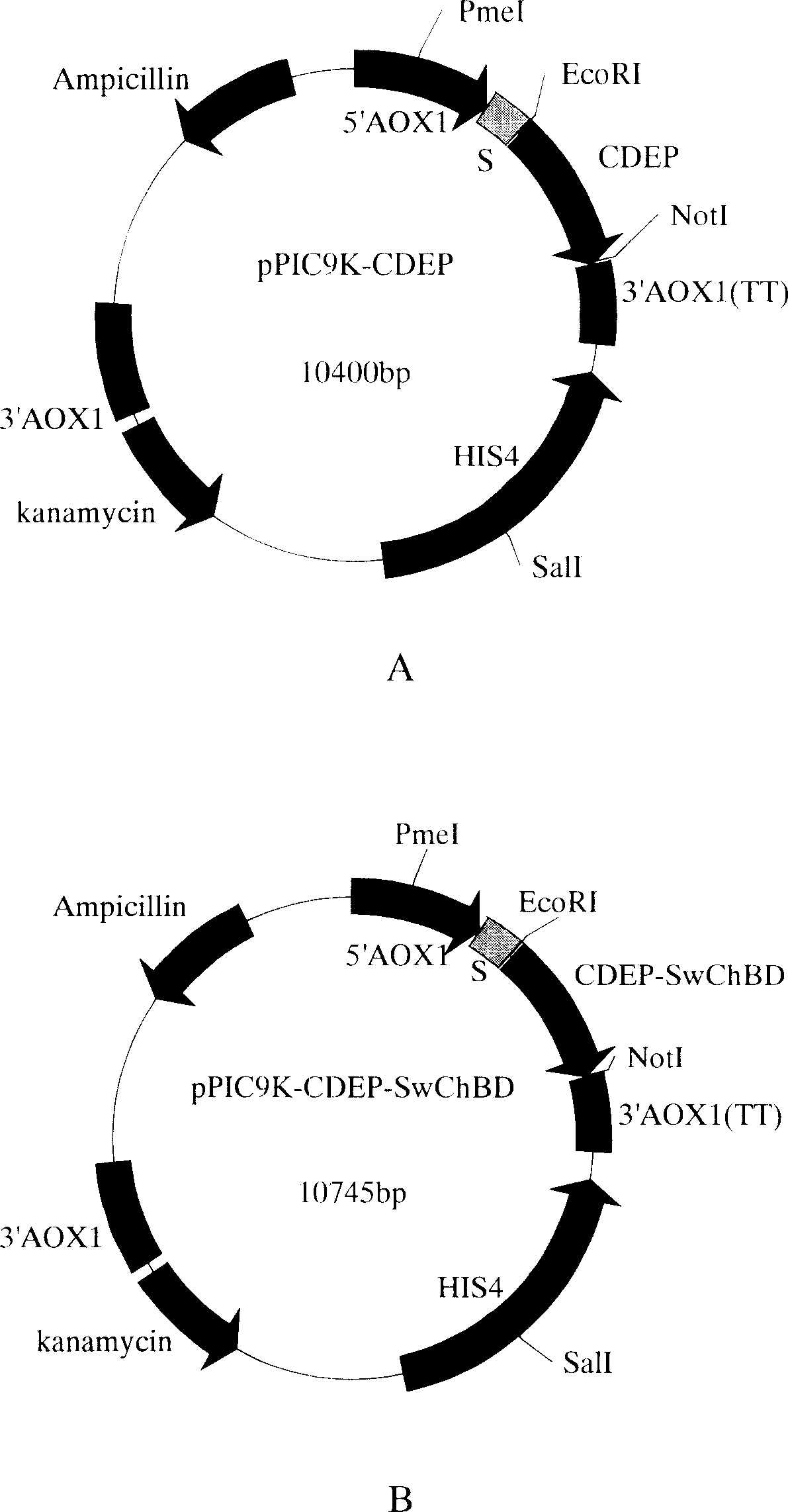
[0066] pMD-CDEP was digested with EcoRI / NotI, and the foreign fragment of about 1 kb was recovered, and cloned into the pPIC9K (Invitrogen Company) vector digested with the same enzyme. It was verified by EcoRI / NotI digestion, and the positive clone was named pPIC9K-CDEP, and its plasmid structure is shown in figure 1 A (The amino acid sequence of the expression product is shown in SEQ ID NO.9).
[0067] 2. Construction of CDEP-SwChBD yeast expression vector
[0068] Digest pMD-hCDEP with EcoRI / XbaI, recover an exogenous fragment of about 1kb, digest pMD-SWChBD with SpeI / NotI, and recover a 340bp exogenous fragment, and clone the two fragments recovered above into EcoRI / NotI enzyme at the same time Cut pPIC9K (Invitrogen) vector. The positive clone identified by EcoRI / NotI digestion was named pPIC9K-CDEP-SwChBD, and its plasmid structure is shown in figure 1 B, the amino acid sequence of the expression product is s...
Embodiment 3
[0079] 1. Construction and transformation of CDEP-1 transformation vector of Beauveria bassiana
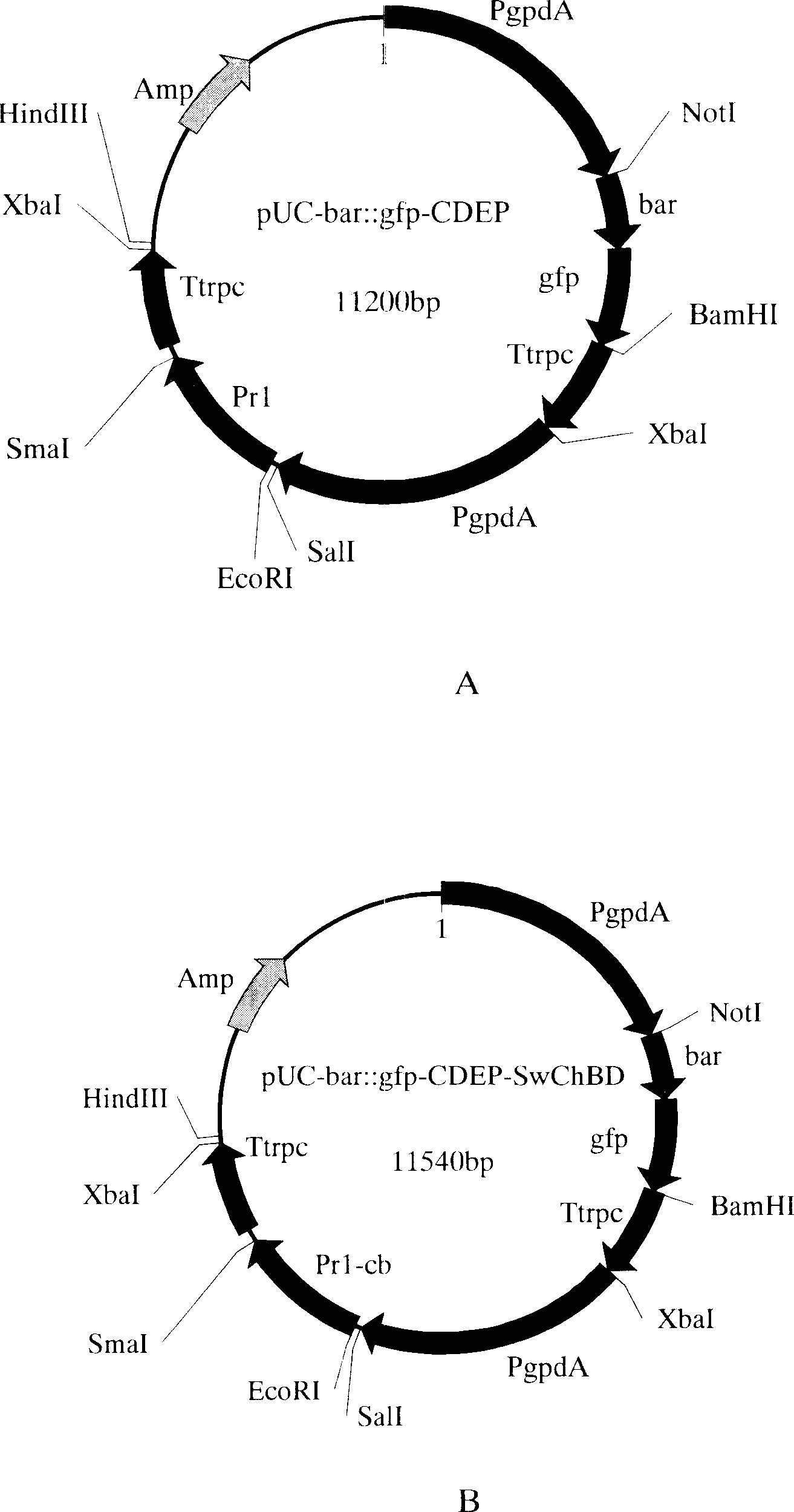
[0080] Digest pMD-spCDEP with EcoRI / SmaI, recover a 1140bp fragment, digest pUC-bar::gfp with SmaI / HindIII, recover the 750bp Trp terminator, and digest pUC-bar::gfp with EcoRI / HindIII , Recover the vector sequence of 10000bp. The three recovered fragments were ligated and transformed into Escherichia coli Dh5α. Validation was performed with EcoRI / HindIII. The positive clone identified by enzyme digestion was named pUC-bar::gfp-CDEP, and its plasmid structure is shown in figure 2 a. The plasmid of the positive clone was extracted and transformed into Beauveria bassiana Bb0062 strain after linearization with HindIII (Beauveria bassiana (Beauveriabassiana) Bb0062 was isolated from the cabbage caterpillar (Pieris rape) infected, preserved in the Southwest Agricultural University Biotechnology Center, It can also be obtained from known sources, such as CCTCCAF93297 or isolated fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com