Method for externally amplifying specific ring type or concatemer nucleic acid
An in vitro amplification, nucleic acid technology, applied in the field of amplifying circular or tandem DNA, can solve the problems of loss of specificity and selectivity, labor, and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
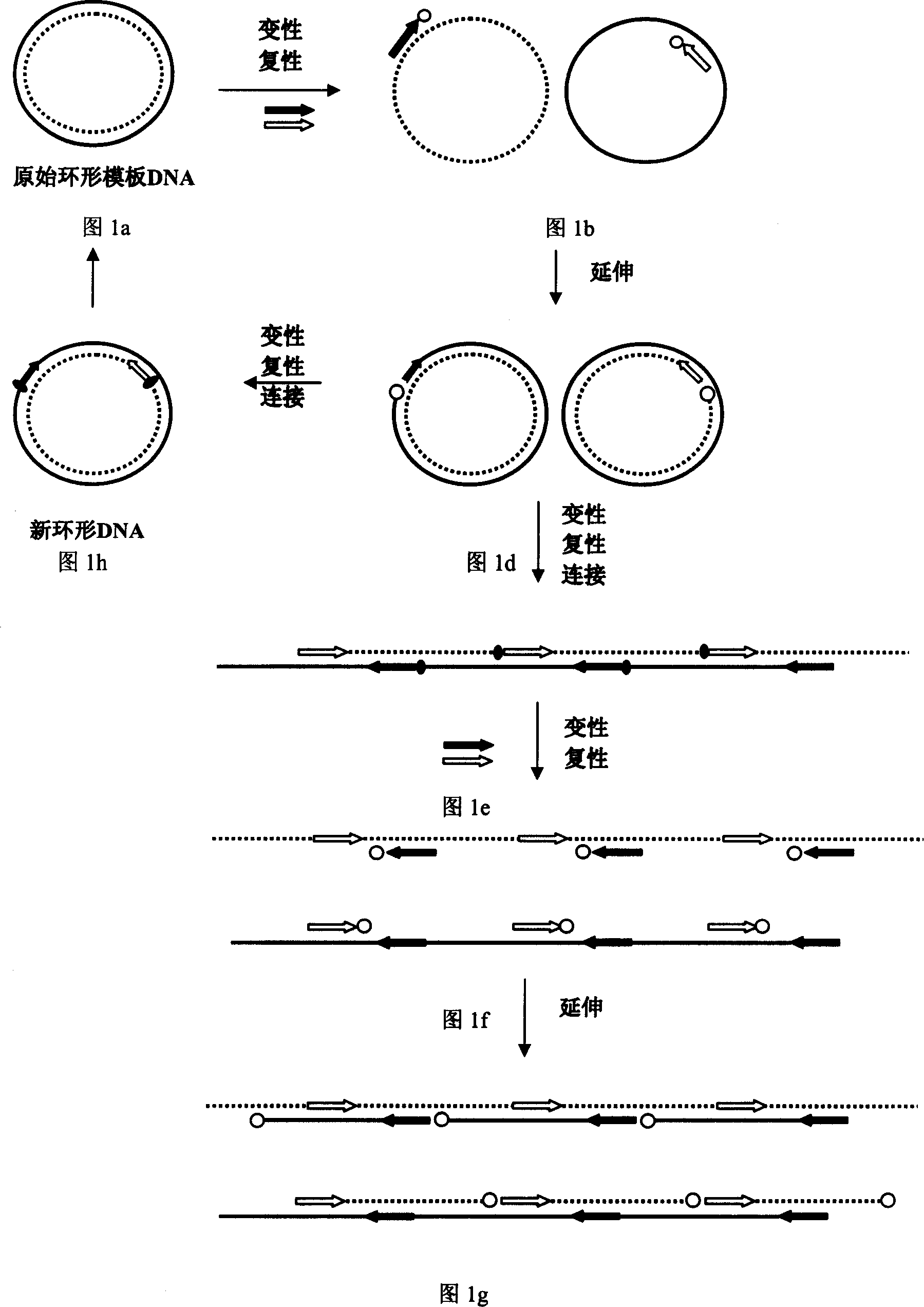
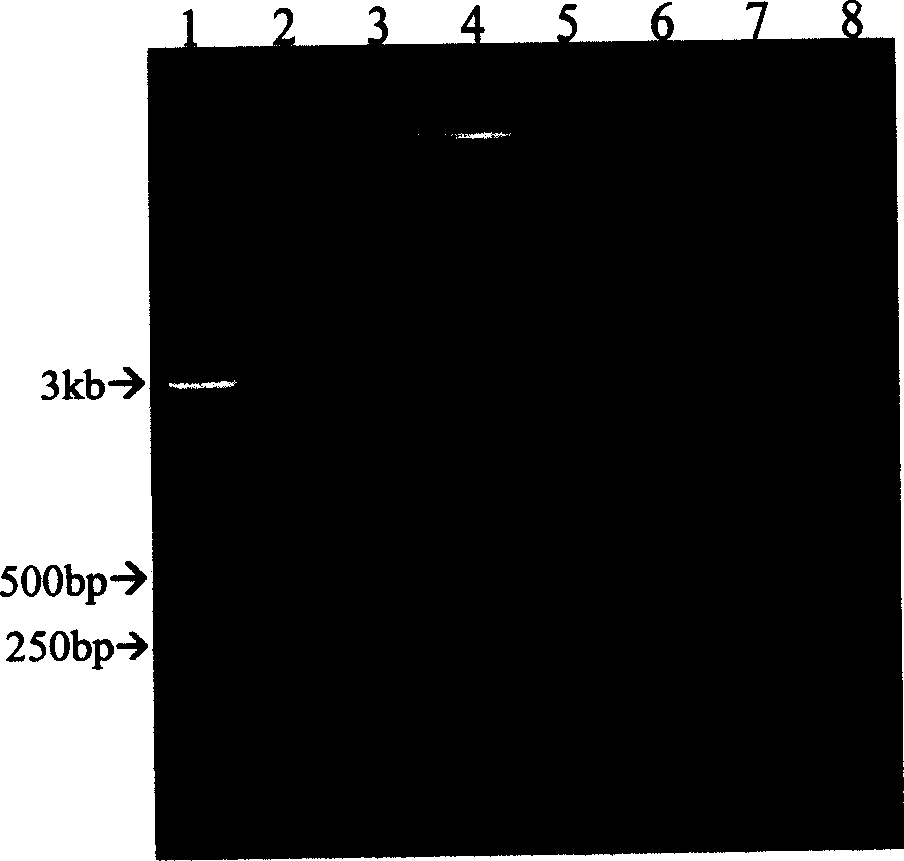
[0066] In order to prove the feasibility of using the LCA method to amplify and produce circular and linear tandem DNA, use the closed circular natural plasmid pUC19 without an insert or with a 300bp to 2kb insert as a template, and use the method described in the present invention to amplify .
[0067] concatenated amplification of closed circular plasmid DNA
[0068] Mix 10 ng of natural closed circular plasmid pUC19 (no insert or 300bp-2kb insert) and 1.0μM M13 forward and reverse primers, add to 50μl reaction system, which contains 0.5xLA-Taq DNA polymerase buffer (TaKaRa), 0.5x Taq DNA Ligase Buffer (20mM Tris-HCl, 25mM Potassium Acetate, 10mM Magnesium Acetate, 1mM NAD + , 10 mM dithiothreitol, 0.1% Triton X-100) and 400 μM of the four mononucleotides. After pre-denaturation at 95°C for 5 minutes, 5 units of LA thermostable DNA polymerase (TaKaRa) and 40 units of thermostable DNA ligase (New England Biolabs) were added to the reaction mixture. Subsequently, 30 cycles ...
specific Embodiment approach 2
[0085] In this embodiment, the plasmid pCMV2-EGFP was amplified by the same method as Embodiment 1, and the 5' phosphorylation primers used were CMV-F and BGH-R. The amplified circular or tandem DNA was digested with EcoRI and then circularized with T4 DNA ligase, transformed into Escherichia coli JM109, and the bacteria expressed green fluorescent protein, which can be directly observed under ultraviolet light.
[0086] The primer sequences used are as follows:
[0087] CMV-F: CGCAAATGGGCGGTAGGCGTG
[0088] BGH-R: TAGAAGGCACAGTCGAGG
specific Embodiment approach 3
[0089] In this embodiment, the plasmid pCMV2-EGFP was amplified by the same method as Embodiment 1, and the 5' phosphorylation primers used were CMV-F and GFP-R. GFP-R contains three random nucleotides, which can be used to introduce mutations within the green fluorescent protein gene. The amplified circular or tandem DNA was digested with EcoR I and then circularized with T4 DNA ligase, transformed into Escherichia coli JM109, and the bacterium expressed the mutated green fluorescent protein, which could be directly observed under ultraviolet light.
[0090] The primer sequences used are as follows:
[0091] CMV-F: CGCAAATGGGCGGTAGGCGTG
[0092] GFP-R: GCGGACTGNNNGCTCAGGTAG
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com