Pyrrolysyl-tRNA synthetase mutant and application thereof
A technology of pyrrolysyl and synthetase, which is applied in the field of enzyme engineering, can solve the problems of low efficiency of unnatural amino acids, achieve the effects of improving introduction efficiency, increasing recognition rate, and facilitating popularization and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Construction of MbPylRS (N311A / C313A) and activity comparison with MmPylRS (N346A / C348A)
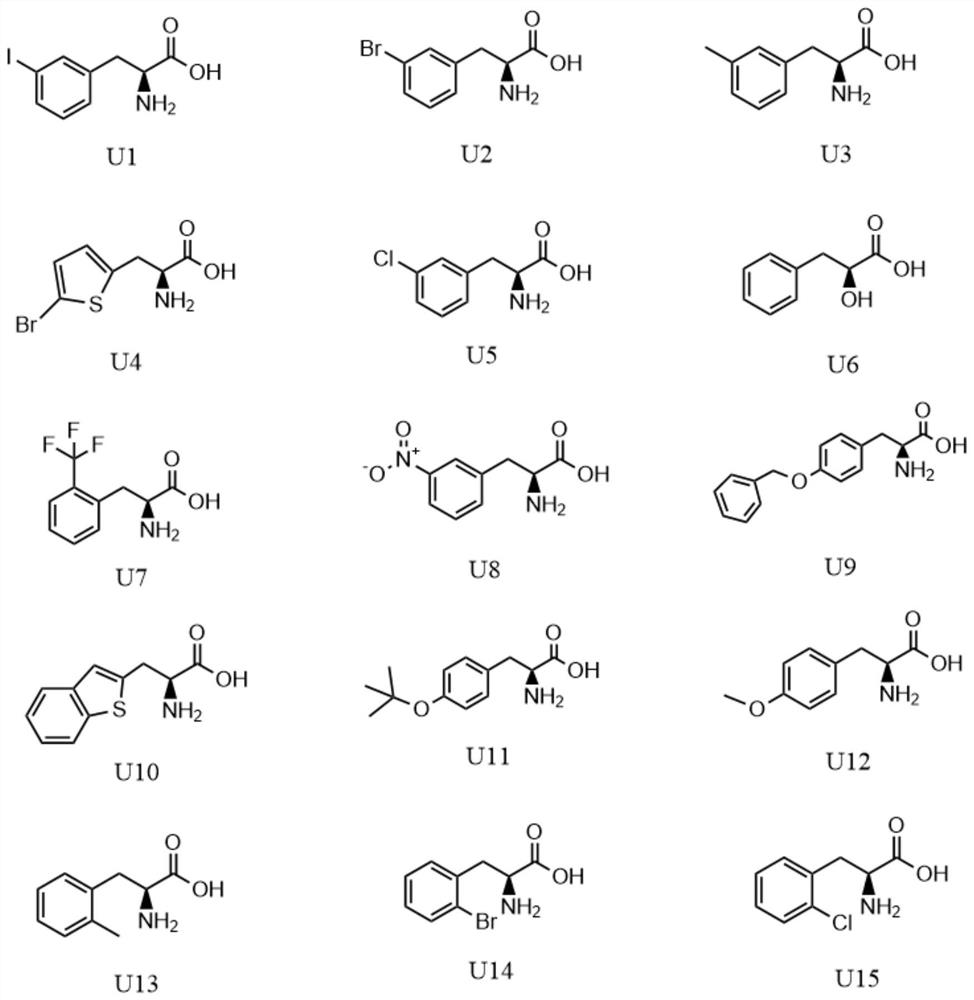
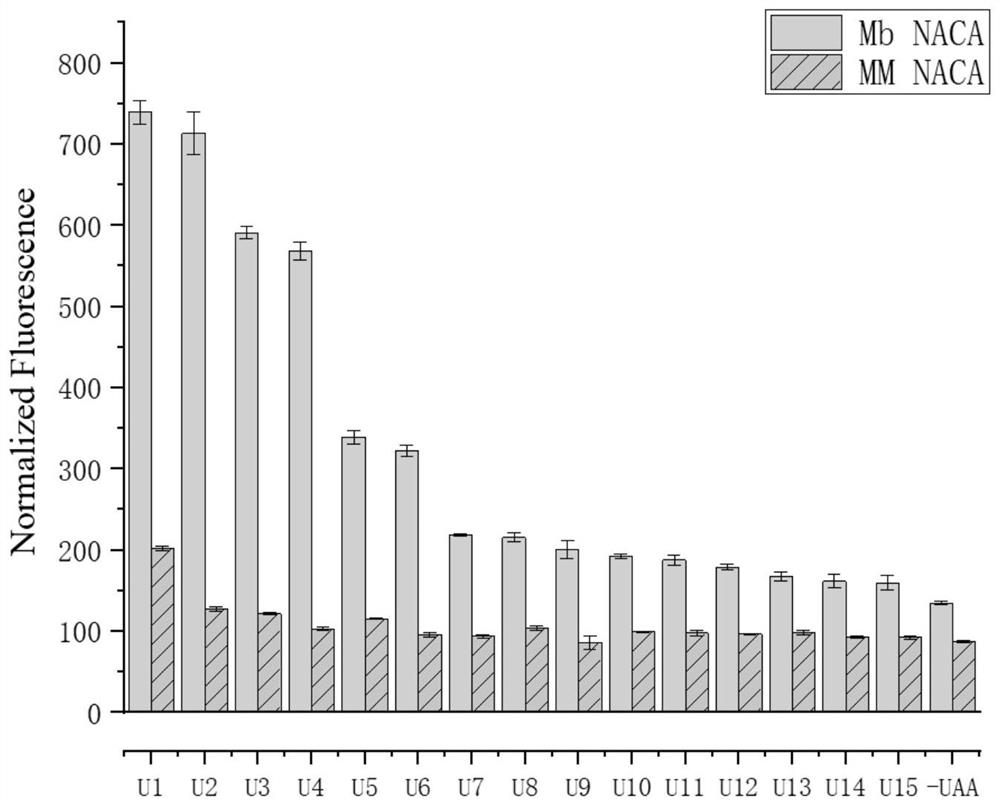
[0035] MmPylRS comprises 454 amino acid residues and has 74% homology with MbPylRS. It is reported that MmPy lRS(N346A / C348A) has high substrate procrastination and can accept more than 40 unnatural amino acids. Based on the sequence comparison between MmPylRS and MbPylRS, the present invention uses site-directed mutagenesis to construct pBK-MbPylRS (N311A / C313A), and detects the expression of green fluorescent protein (superfolder green fluorescent protein, sf GFP) on 15 different benzene The alanine analog unnatural amino acid was tested for introducing efficiency. The expression vector pGFP2TAG was obtained by using the pGFP vector as a template to mutate the serine triplet base sequence at the second site of the GFP protein into TAG by PCR.
[0036] The activity detection method is as follows:
[0037] 1. Plasmids pBK-MbPylRS (N311A / C313A) and pBK-MmPylRS (N346A / C3...
Embodiment 2
[0045] Example 2 Construction and activity detection of MbPylRS (V31I, T56P, A100E, N311A, C313A) mutants
[0046] PylRS mainly contains two functional domains, the C-terminal catalytic domain consisting of about 250 amino acid residues, and the N-terminal tRNA binding domain consisting of about 140 amino acid residues. The present invention uses MbPylRS (N311A / C313A) as a template to introduce a series of mutations at the N-terminal of PylRS to further improve its catalytic efficiency.
[0047] 1. Construction of single point mutants
[0048] Design mutation primers according to the nucleotide sequence of MbPylRS (SEQ ID NO.2), using fast PCR technology, using the recombinant vector pBK-MbPylRS (N311A / C313A) as a template;
[0049] (1) A single mutation is introduced at position 31 of the amino acid sequence of MbPylRS, and the primers are:
[0050] Forward primer 1: 5'-catgaa att agccgcagcaaaatctatattga-3' (the underline is the mutated base)
[0051] Reverse primer 2: 5...
Embodiment 3
[0080] Example 3 Expression and purification of GFP protein introducing unnatural amino acid
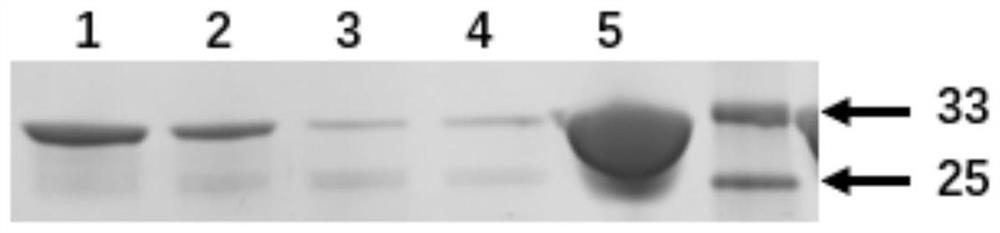
[0081] The present invention utilizes MbPylRS (V31I / T56P / A100E / N311A / C313A) to introduce an unnatural amino acid into the second amino acid site of the GFP protein, and induces the expression to purify the GFP protein introduced with the unnatural amino acid ( image 3 ), providing broad prospects for expanding the application of unnatural amino acids in the field of enzyme engineering.
[0082] The expression purification method is as follows:
[0083] 1. Plasmids pBK-MbPylRS (V31I / T56P / A100E / N311A / C313A) and pGFP2TAG were co-transformed into E. coli transformation-competent DH10B; +50μg / mL kanamycin resistance plate; 37 ℃ inverted culture overnight;
[0084] 2. Pick a single colony into 5mL LB medium containing KT resistance, culture overnight at 37°C and 220rpm;
[0085] 3. The next day, transfer 1% transfer volume to 400mL LB liquid medium, culture at 37°C 220rpm for 3h, add a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com