Method and system for detecting macro virus group in sample
A macro virus, in-sample technology, applied in the field of macrovirome detection in samples, can solve the problems of low virus sequence integrity, low virus sequence quality, short contigs, etc., to improve the identification sensitivity and accuracy, and improve the sensitivity. and accuracy, wide range of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Embodiment 1 detects the method for the macrovirus group in the sample
[0067] 1. Sample processing
[0068] Divide the samples into 2 parts, and carry out the experimental processing on the second-generation and third-generation sequencing platforms respectively, as follows:
[0069] 1. Next generation sequencing
[0070] 1) Firstly, total DNA was extracted by phenol-chloroform extraction. After the nucleic acid was extracted, two instruments, Thermo NanoDrop 2000 and Qubit, were used to measure the concentration and quality of the nucleic acid respectively;
[0071] 2) DNA fragments are purified and eluted by the magnetic bead method, and the ends of the fragments are repaired;
[0072] 3) Add adapters and amplify the DNA fragments to construct a sequencing library;
[0073] 4) Use the Illumina Novaseq platform for metagenomic sequencing of PE150. After the sequencing is off the machine, perform data format conversion and index splitting on the original bam file ...
Embodiment 2
[0113] Embodiment 2 detects the system of the macrovirus group in the sample
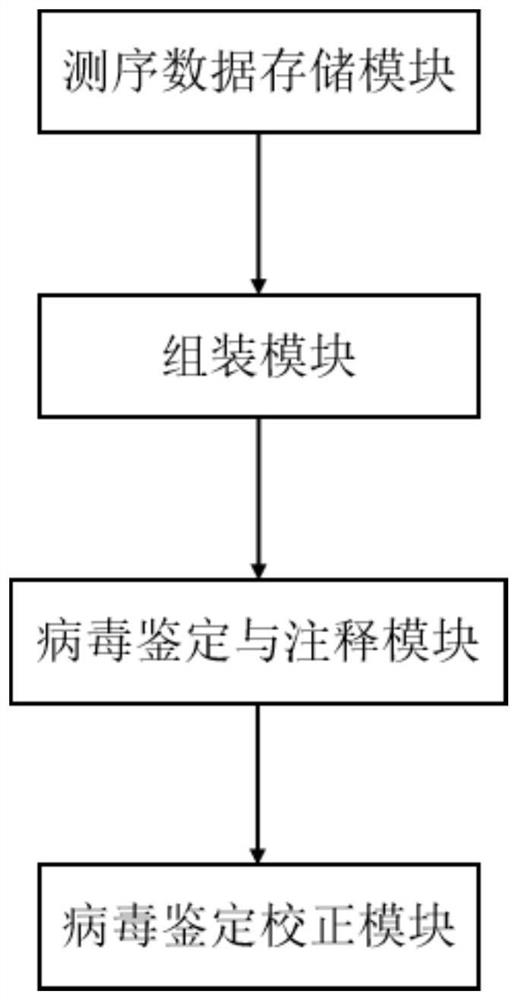
[0114] This embodiment provides a system for implementing the method for detecting macroviruses in samples in Embodiment 1, such as figure 1 shown, including:
[0115] The sequencing data storage module is used to obtain and store the second-generation sequencing data and the third-generation sequencing data of the samples to be tested;
[0116] The assembly module is connected with the sequencing data storage module, and is used for mutual correction and mixed assembly of the second-generation sequencing data and the third-generation sequencing data to obtain mixed assembly contigs, and is used to separately assemble the third-generation sequencing data to obtain Nanopore contigs;
[0117] The virus identification and annotation module, connected with the assembly module, is used to compare and annotate the mixed assembly contigs, obtain candidate virus contigs and non-virus contigs, and perform s...
Embodiment 3
[0120] Example 3 Human stool samples were subjected to metagenomic sequencing and analysis
[0121] 1. Sample processing
[0122] After the sample was divided into two parts according to the method of Example 1, the second-generation metagenomic sequencing and the third-generation metagenomic sequencing were performed respectively.
[0123] 2. Sequencing data analysis
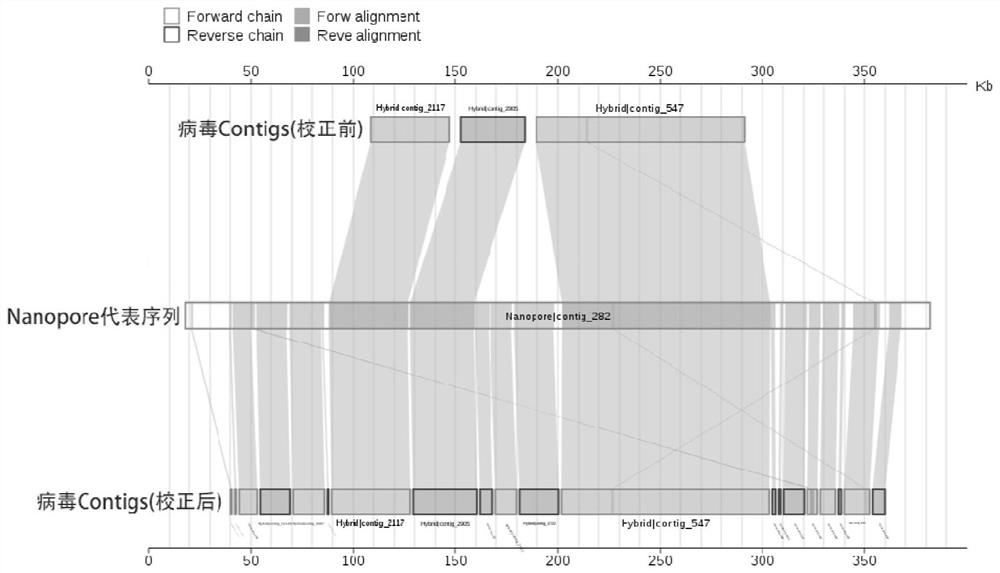
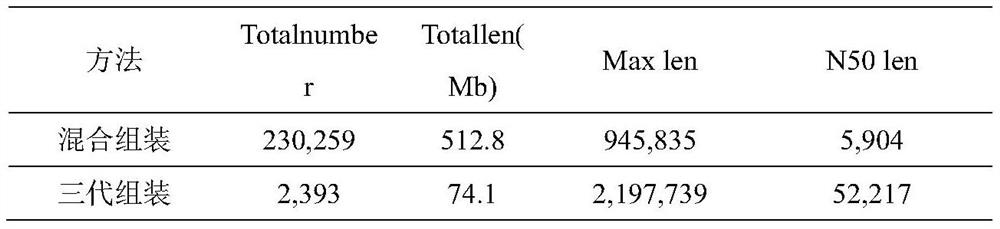
[0124] According to the method of Example 1, the second-generation sequencing data volume of the sample is about 10Gb bases, and the third-generation sequencing data volume is about 6Gb bases. After removing the human host sequence, the remaining 9.2Gb of the second-generation data and 5.64Gb of the third-generation data. The mixed assembly of the second-generation and third-generation quality control data obtained 230,000 contigs, with a total of 512.8Mb bases; and the separate assembly of the third-generation quality control data obtained more than 2,000 contigs, with a total of 74.1Mb bases (Table 1).
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com