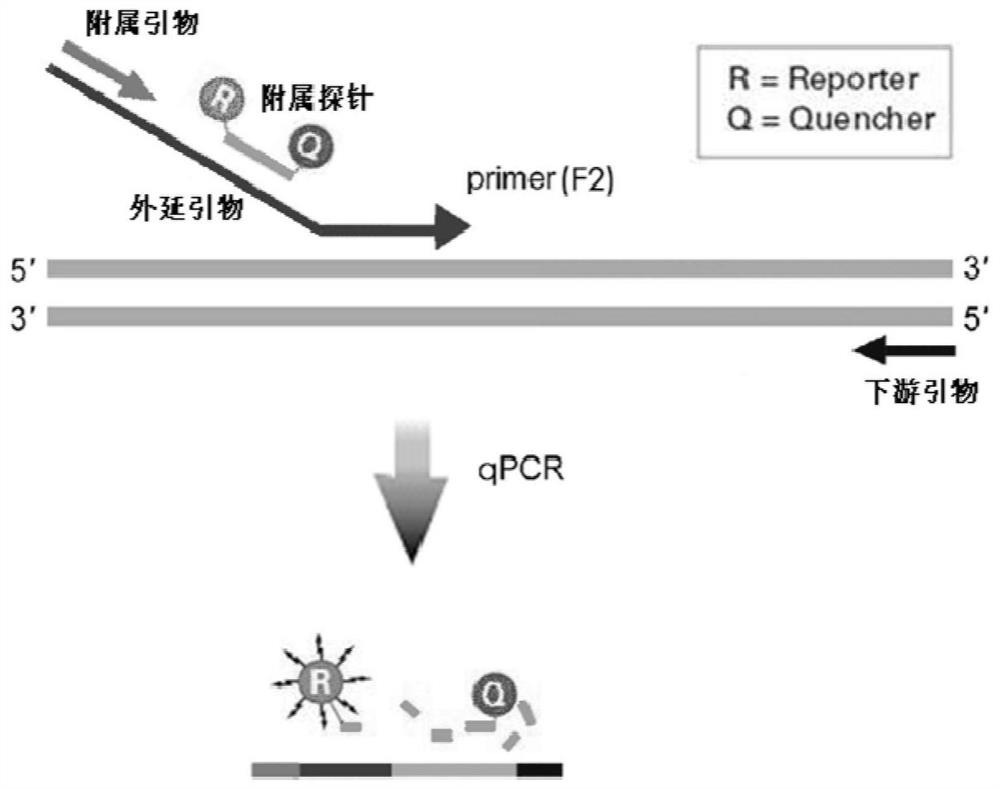
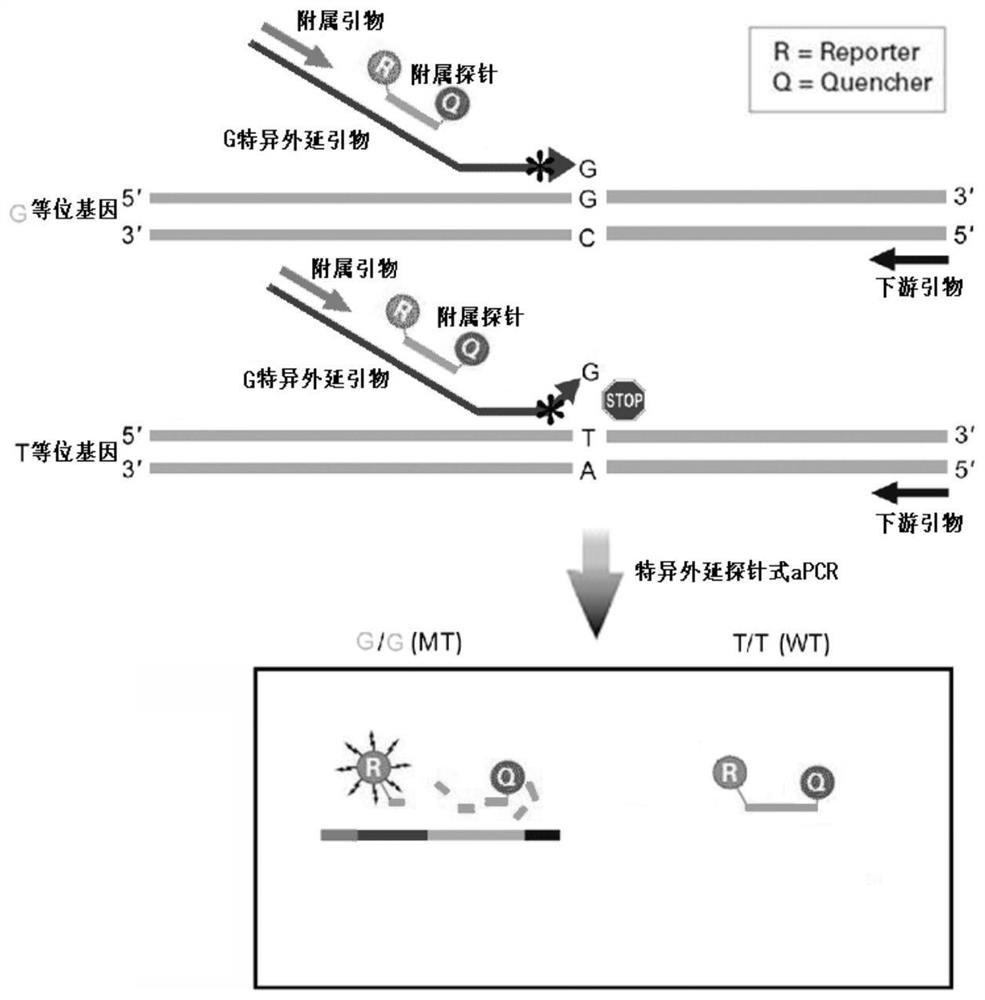
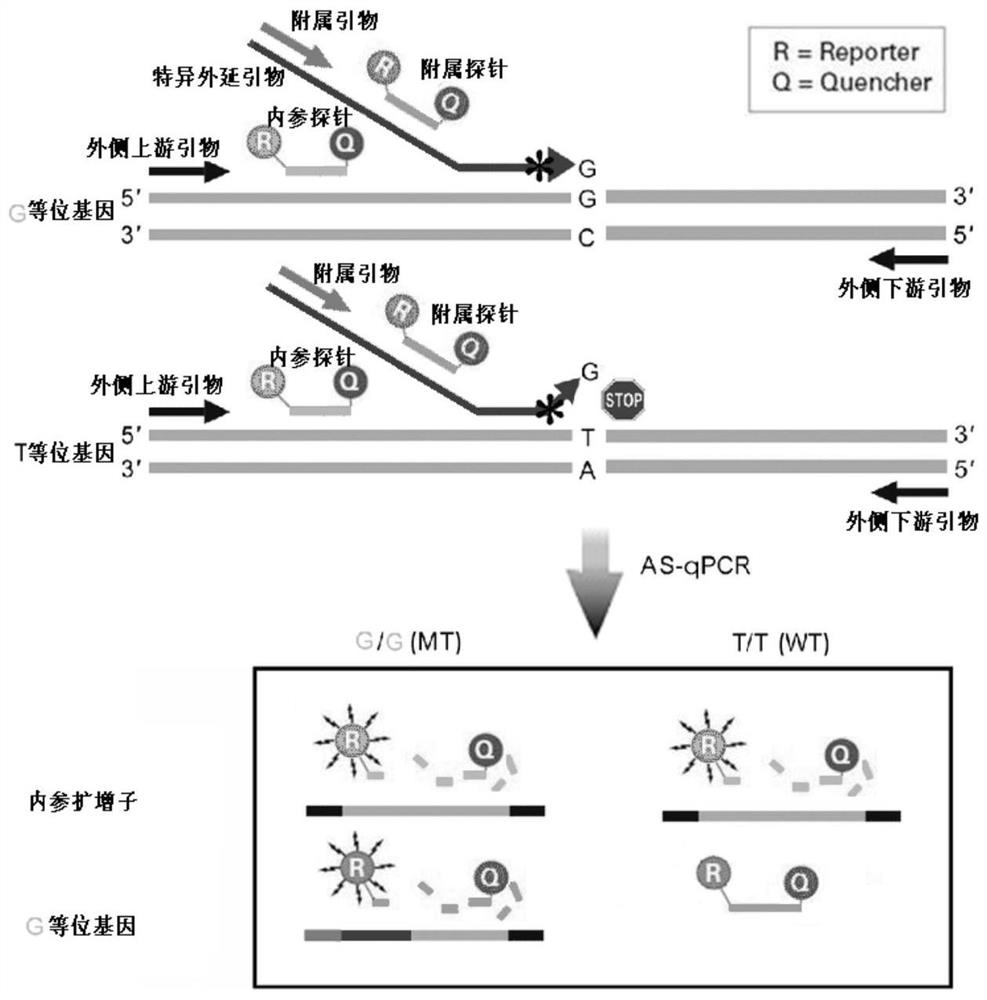
Epitaxial probe type qPCR detection method for SNV
A detection method and probe-based technology, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. It can solve the problems of decreased binding efficiency, decreased detection sensitivity and specificity, and achieve high sensitivity and specificity, cost-reducing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Detection of New Coronavirus N501Y
[0058] We intend to develop a new coronavirus N501Y site detection kit based on the unidirectional epitaxial probe qPCR detection system to prove the effectiveness of the unidirectional epitaxial probe qPCR detection system.
[0059] 1 Materials and methods
[0060] 1.1 Preparation of N501Y mutant and wild-type plasmids
[0061] According to the gene sequence of SARS-CoV-2 VOC 202012 / 01" or "B.1.1.7." entered by GenBank, select the 1398th to 1588th sites in the spike protein gene sequence (its position in the full-length sequence 22935 to 23125), the wild-type sequence and the N501Y mutant sequence were synthesized by General Biosystems (Anhui) Co., Ltd. by splicing primers, and inserted into the EcoRV blunt-end restriction site of the puc57 plasmid. The plasmid was transformed and extracted Finally, the wild-type plasmid and the mutant plasmid were diluted with double distilled water to 1×10 to the 10th power copies / uL, ...
Embodiment 2
[0099] The detection of embodiment two multiple SNPs
[0100] We plan to develop c.-3279T>G and c.1091C>T detection methods on UGT1A1 based on the bidirectional epitaxial probe qPCR detection system to prove the effectiveness of the bidirectional epitaxial probe qPCR detection system in SNP detection and the fluorescent probe Multiplexability in detecting different SNPs.
[0101] 1 Materials and methods
[0102] 1.1 Subjects under inspection
[0103] Peripheral blood samples from 22 neonates, aged 0-1 years, all came from the Neonatology Department of Shanghai Children's Hospital. The sample acquisition has obtained the informed consent of the legal representative or family members.
[0104] 1.2 Sanger sequencing
[0105] By amplifying the UGT1A1 exon, c.-3279T>G attachment region, and then performing Sanger sequencing, the variation of the subject's UGT1A1 gene was detected.
[0106] 1.3 Design of primers and probes for two-way epitaxial probe qPCR detection system for d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com