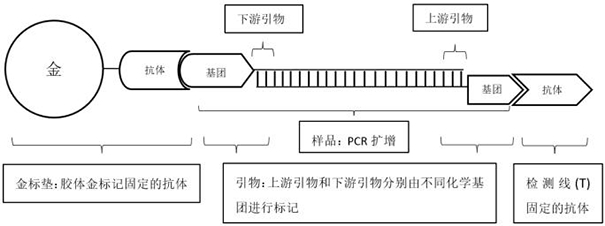
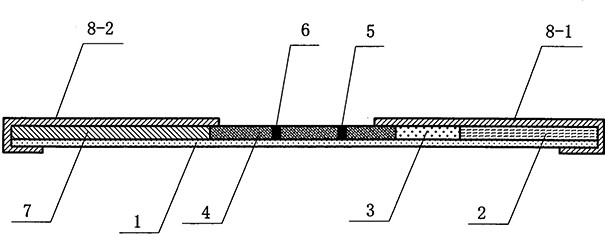
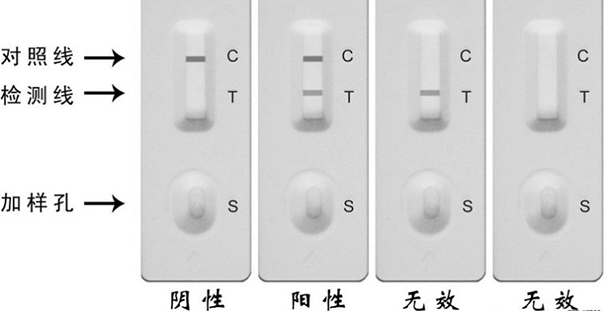
Amplification primer composition and kit for detecting new coronavirus
A coronavirus and amplification primer technology, applied in the biological field, can solve the problems of high operation requirements of medical personnel, high operator requirements, and large investment in equipment costs, avoiding false positives, increasing annealing temperature, and high amplification efficiency. and specific effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] 1. Primer design for PCR of 2019-nCoV
[0089] Referring to the 2019-nCoV genome sequence included in GenBank, select the conserved regions of ORF1ab gene and N gene of 2019-nCoV, and use ABI PrimerExpress 3.0 PCR primer design software to design and synthesize primers. The final sequences of each primer are as follows.
[0090]
[0091] 2. Locked Nucleic Acid Modified PCR Primers
[0092] The primer modification used in this embodiment is: each upstream primer and downstream primer has four bases modified by locked nucleic acid (+N bases are modified by locked nucleic acid).
[0093] 2.1 2019-nCoV-ORF1ab Locked Nucleic Acid Modified Primers
[0094] 2.1.1 2019-nCoV-ORF1ab four base-locked nucleic acid modified primers (see Table 2 above for details)
[0095] F: 5'-AATGT +T AATGC +A CTTTT +A TCTAC +T -3'
[0096] R: 5'-AACCG +T TCAA +T CATA +A GTGT +A -3'
[0097] 2.1.2 2019-nCoV-ORF1ab zero base-locked nucleic acid modified primers
[0098] F: 5'-AATGTTAA...
Embodiment 2201
[0181] Example 2 Detection Kit for 2019-nCoV PCR Amplification Combined with Nucleic Acid Capture Gold Standard Test Strip
[0182] Locked nucleic acid modified primers 2019-nCoV-ORF1ab-F (10 μM) 40 μL for PCR detection of 2019 novel coronavirus (2019-nCoV) nucleic acid, 2019-nCoV-ORF1ab-R (10 μM) 40 μL, 2019-nCoV- 40 μL of N-F (10 μM), 40 μL of 2019-nCoV-N-R (10 μM), 250 μL of master mix, 250 μL of 2mM deoxyribonucleotide, Mn 2+ 60 μL, 15 μL of amplification enzyme, 20 μL of 2019-nCoV plasmid DNA (30ng / μL), 1.5 mL of ultrapure water, 5 mL of loading sample diluent, and 48 finished test strips were packaged together to obtain the 2019 novel coronavirus (2019-nCoV ) PCR product nucleic acid capture gold standard test strip detection kit.
[0183] In summary, the kit and detection method provided by the present invention have the characteristics of high sensitivity, strong specificity, and good stability, and can be used for the rapid detection of 2019 novel coronavirus pneumo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com