Molecular marker for predicting Meloidogyne incognita resistance of Prunus sogdiana
A M. incognita, molecular marker technology, applied in biochemical equipment and methods, microbial assay/test, DNA/RNA fragment, etc. Problems such as the breeding process of new varieties of cherry plum resistant to southern root-knot nematode
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Preparation of Molecular Markers for Predicting Resistance to Meloidogyne incognita Xinjiang Wild Cherry Plum
[0046] 1. Breeding of wild cherry plums and plants resistant to root-knot nematode incognita. Taking the wild cherry plum in Tianshan Mountains of Xinjiang as the original resource, the seeds were collected, and the seedling population was obtained by sowing. Multi-year identification of resistance of seedling populations to Meloidogyne incognita. The individual plants 'K-323', 'K-325', 'K-510' resistant to M. incognita and 'G-319', 'G-333', 'G-348' susceptible to M. incognita were obtained '. Grafting compatibility tests on single plants with high resistance to root-knot nematode in Xinjiang showed that wild cherry plums with high resistance to root-knot nematode in Xinjiang had good grafting compatibility with stone fruit trees such as peaches, plums, and apricots. It shows that the resistant rootstock provided by the application can be widely u...
Embodiment 2
[0076] Example 2 Method for Predicting Meloidogyne incognita in Xinjiang Wild Cherry Plum Using Molecular Markers
[0077] This example provides the application method of molecular markers of Xinjiang wild cherry plums resistant to root-knot nematode incognita. The Xinjiang wild cherry plums used in this example are 'K-325', 'K-510' and individual plants 'G-319', 'G-333', 'G-348', 'G-402 susceptible to Meloidogyne incognita '.
[0078] 1. Using the same method as in step 2 of Example 1, carry out the extraction of Xinjiang wild cherry plum genome cDNA.
[0079] 2. Using the oligonucleotide primers SEQ ID No.1-2 designed in Example 1 as PCR amplification primers.
[0080] 3. PCR polymerase chain reaction amplification and agarose gel electrophoresis detection:
[0081] (1) PCR reaction system: 2×M5HiPer plus Taq HiFi PCR Mix 12.5μl; cDNA template 2.5μl; SEQ ID No.1 1.25μl; SEQ ID No.2 1.25μl; deionized water 7.5μl;
[0082] (2) PCR reaction conditions: pre-denaturation at 9...
Embodiment 3
[0085] Example 3 Molecular Marker Prediction Accuracy of Xinjiang Cherry Plum's Resistance to Meloidogyne incognita and Grafting Affinity
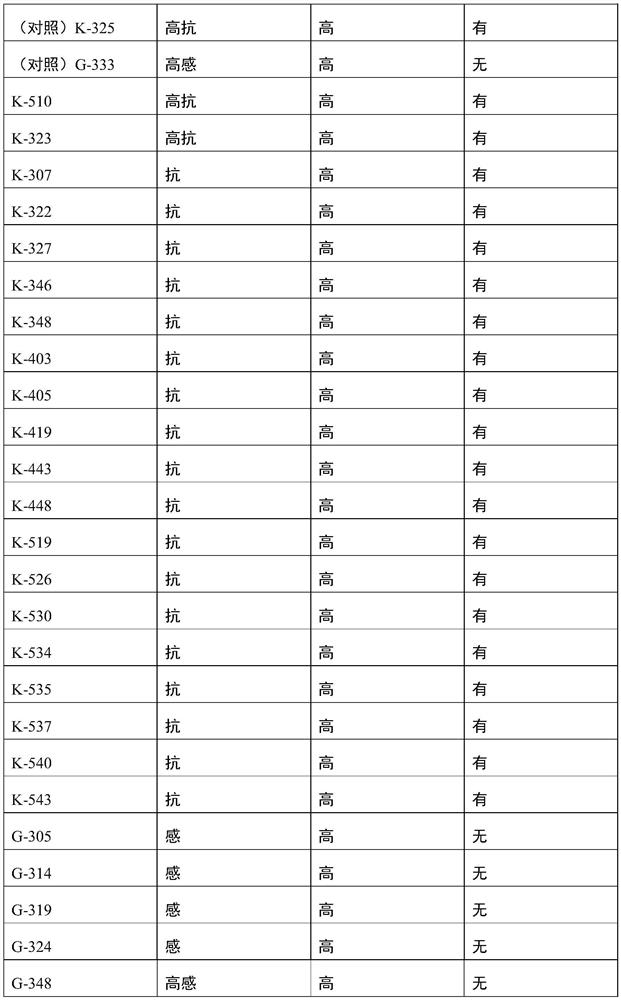
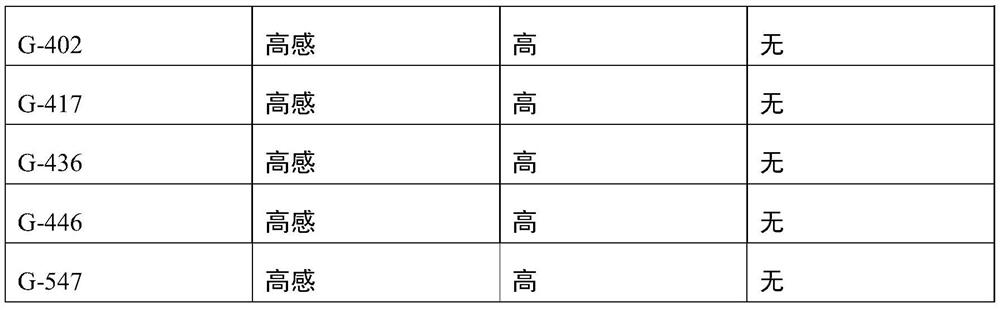
[0086] EXAMPLES The primers (SEQ ID No.1-2) designed in Example 1 were used to predict the resistance to root-knot worm and grafting affinity of 30 Xinjiang cherry plums, and the results are shown in Table 1.
[0087] Table 1 The results of prediction using SEQ ID No.1-2
[0088]
[0089]
[0090]
[0091] As can be seen from the results in Table 1, using the primer pair shown in SEQ ID No.1-2, carry out the resistance prediction of root-knotworm for 30 strains of Xinjiang wild cherry plum, and the prediction accuracy rate reaches 100%; Confirm that the molecule provided by the present invention The markers are significantly correlated with the resistance to root-knot worm incognita. The primer pair provided by the invention can not only be used to predict the resistance to root-knot worm in Xinjiang wild cherry plums, but also can ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com