SSR (Simple Sequence Repeat) marker for simultaneously identifying sterile cytoplasm of gossypium harknessii and gossypium hirsutum
A Hackney West cotton and cytoplasmic technology, applied in the field of SSR labeling, can solve the problem of inability to identify two different sterile cytoplasmic substances, achieve the effects of improving accuracy and efficiency, wide application range, and saving manpower
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
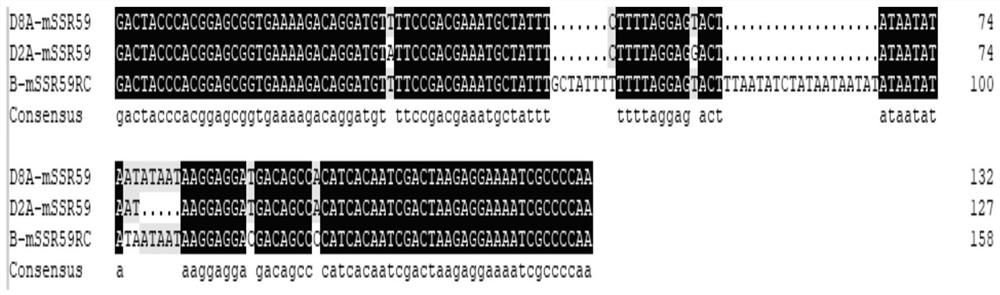
[0026] Using the leaves of cotton sterile lines and maintainer lines containing the male sterile cytoplasm of Harknessy cotton (CMS-D2) and cotton trilobes (CMS-D8) as research materials, 64 pairs of mitochondrial SSR markers (according to ncbi The design of the mitochondrial genome sequence of the Hackney cotton sterile line and maintainer line published in the Internet, the mitochondrial genome sequence number is JX536494.1, JX065074.1) screening, a total of 8 can distinguish the sterile line and the maintainer line Mitochondrial SSR marker, a mitochondrial SSR marker (named mSSR59) that can distinguish CMS-D2 from CMS-D8.
[0027] The PCR product of mSSR59 was purified, connected to the T vector, and sequenced in E. coli to obtain a PCR product of CMS-D2 (D2A-mSSR59) of 127bp, a PCR product of CMS-D8 (D8A-mSSR59) of 132bp, and a product of the maintainer line (B -mSSR59) is 158 bp. The PCR product of CMS-D2 is 5bp less "ATAAT" than the PCR product of CMS-D8, and the two ar...
Embodiment 2
[0031] (1) DNA extraction
[0032] The seeds of the cotton sterile line and maintainer line of Hackney West cotton and three-lobed cotton male sterile cytoplasm, and the cotton sterile line, maintainer line and F 1 Mitochondrial DNA was extracted using the CTAB method.
[0033] (2) PCR amplification
[0034] Using the above-mentioned mSSR59 molecular marker primer pair, the extracted DNA was amplified by Taq (5U / ul, Quanshijin Company) system. The PCR amplification system was as follows: 20μL reaction system, containing 10×Buffer (containing Mg 2+ ) 2 μL, dNTP (10 mM) 0.4 μL, Taq enzyme 0.2 μL, upstream and downstream primers (10 μM) each 0.8 μL, DNA template (300~500ng / μL) 0.5 μL and ddH 2 O15.3 μL.
[0035] PCR amplification program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 10 s, 34 cycles; extension at 72°C for 5 min, storage at 4°C.
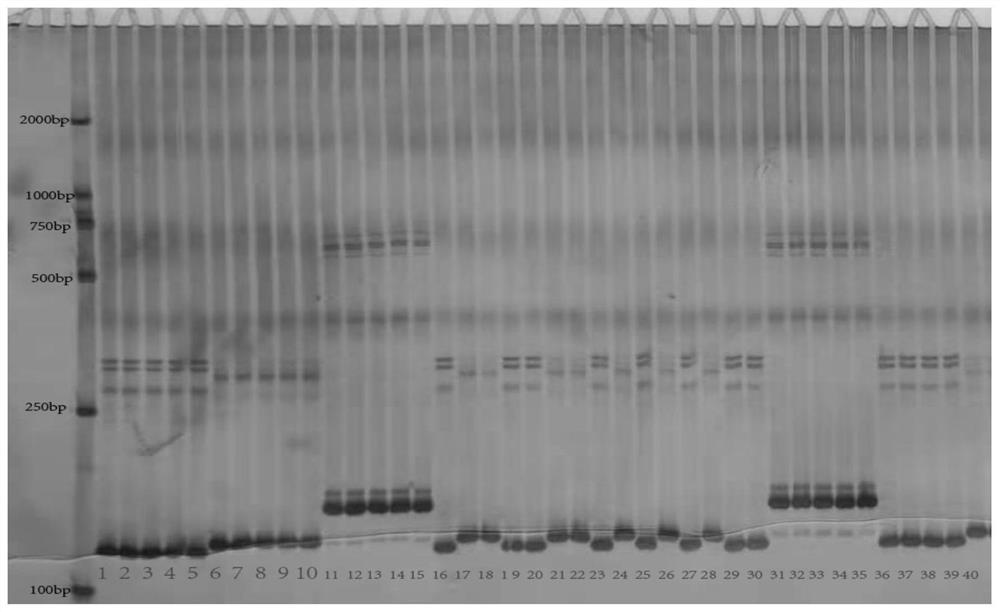
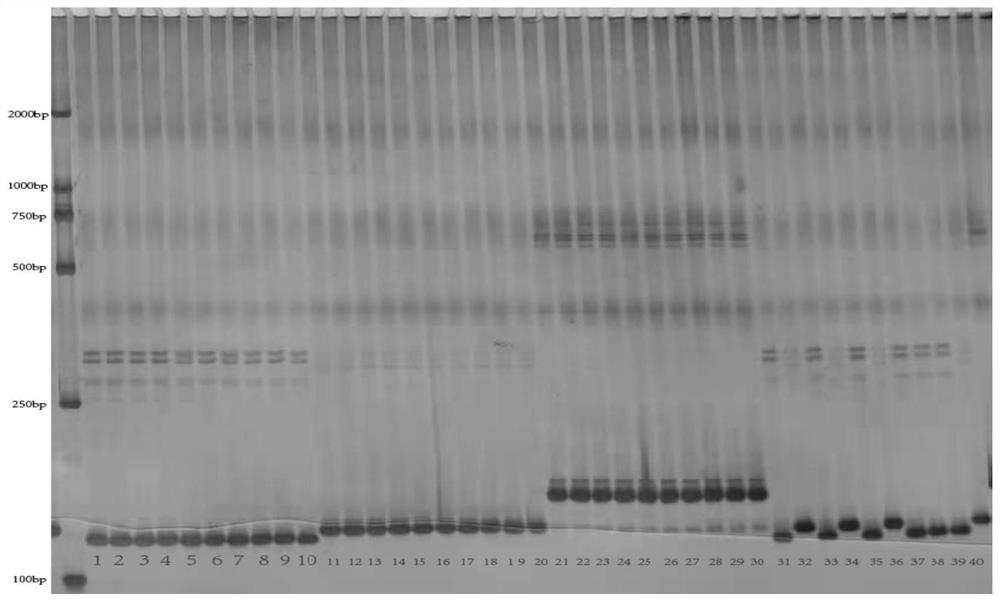
[0036] (3) Polyacrylamide gel electrophoresis
[0037] Aft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com