Colorectal cancer biomarker as well as screening method and application thereof
A biomarker and colorectal cancer technology, applied in the field of colorectal cancer biomarkers and its screening, can solve the problems of low penetration rate and complicated preparation process, and achieve high sensitivity and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Establishment of intestinal microbial marker model for colorectal cancer
[0057] 1. DNA extraction of intestinal bacteria
[0058] Stool samples from 45 colorectal cancer patients and 46 healthy people were collected for screening intestinal biomarkers of colorectal cancer. The specific steps are as follows:
[0059] 1: Aliquot the above stool samples and freeze them at -80°C;
[0060] 2: Use the Surbiopure Fecal Nucleic Acid Extraction Kit (Guangzhou Saibai Pure Biotechnology Co., Ltd.) to extract the DNA of fecal bacteria from colorectal cancer patients and healthy people described in 1. The specific steps are as follows:
[0061] S1. Get 0.25g stool sample (set a blank control (ddH 2 O) Add the positive control (a nucleic acid with known microbial composition and abundance (ZymoBIOMICS Microbial Community Standard; Cat. No. D6300)) to the Drybeads Tube, add 900 μl S1-Lysis Enhancer, vortex thoroughly, and mix well.
Embodiment 2
[0102] Methylation detection of ADHFE1 gene in colorectal cancer
[0103] 1. Primer design
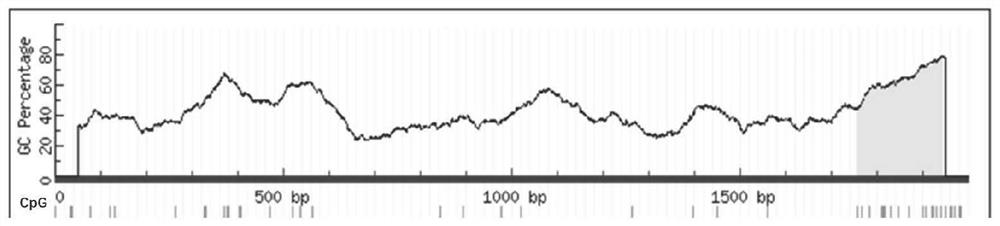
[0104] 1. Using ACTB as an internal reference, design primers for fluorescence quantitative detection according to the position of the CpG island of ADHFE1. The position of the CpG island is as follows: figure 1 Shown; ADHFE1 gene detection primers are as follows:
[0105] ADHFE1-PF: 5'-AGGCCTACGGAGCAGTTA-3' (SEQ ID NO.3);
[0106] ADHFE1-PR: 5'-CCACGGAGTAAGAACCAGAAC-3' (SEQ ID NO.4);
[0107] ADHFE1-NF:5'-TTTGGTTTGTTGGTGAGTAGAG-3' (SEQ ID NO.5);
[0108] ADHFE1-NR: 5'-ACACACCAATTAACAACTCCA-3' (SEQ ID NO. 6).
[0109] 2. Extraction of host DNA
[0110] Fecal samples from 45 patients with colorectal cancer and 46 healthy people were collected respectively, and the samples were subpackaged and frozen at -80°C (same as in Example 1). The host DNA of the above samples was extracted with a blood tissue DNA extraction kit (Tiangen Biochemical Technology Co., Ltd.), and the DNA was stor...
Embodiment 3
[0143] Methylation detection of SDC2 gene in colorectal cancer
[0144] 1. Primer design
[0145] 1. Using ACTB as an internal reference, design fluorescent quantitative detection primers according to the position of the CpG island of SDC2. The position of the CpG island is as follows: figure 2 Shown; SDC2 gene detection primers are as follows:
[0146] SDC2-PF: 5'-TCCTCGGAGCTGCCAATC-3' (SEQ ID NO. 7);
[0147] SDC2-PR: 5'-AGGCGGCAGTGTGACTC-3' (SEQ ID NO.8);
[0148] SDC2-NF: 5'-TTTGGTTTGTTGGTGAGTAGAG-3' (SEQ ID NO.9);
[0149] SDC2-NR: 5'-ACACACCAATTAACAACTCCA-3' (SEQ ID NO. 10).
[0150] 2. The extraction, methylation treatment and methylation detection steps of the host DNA are the same as in Example 2.
[0151] 3. Results
[0152] The methylation detection results of SDC2 are as follows: Image 6 Shown: When the positive control primer Tm value=79.5 is used as the critical value, the correct rate of CRC samples is 84.44%, and the correct rate of healthy samples is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com