Reagent and method for detecting microRNA based on click chemical connection and hairpin stacking assembly
A technology of click chemistry and reagents, which is applied in biochemical equipment and methods, microbiological determination/inspection, etc. It can solve the problems of complex detection steps and achieve the effects of easy control, excellent performance and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, synthetic probe sequence
[0039] Referring to Table 1, the probes and other nucleic acid sequences used in the present invention were synthesized.
[0040] All nucleic acid sequences used in the present invention in table 1
[0041]
[0042]
[0043] Note: The underline in the table indicates the stem sequence of the DNA hairpin probe, and the italic indicates the loop sequence. The gray font indicates the mutated base site of let-7a.
Embodiment 2
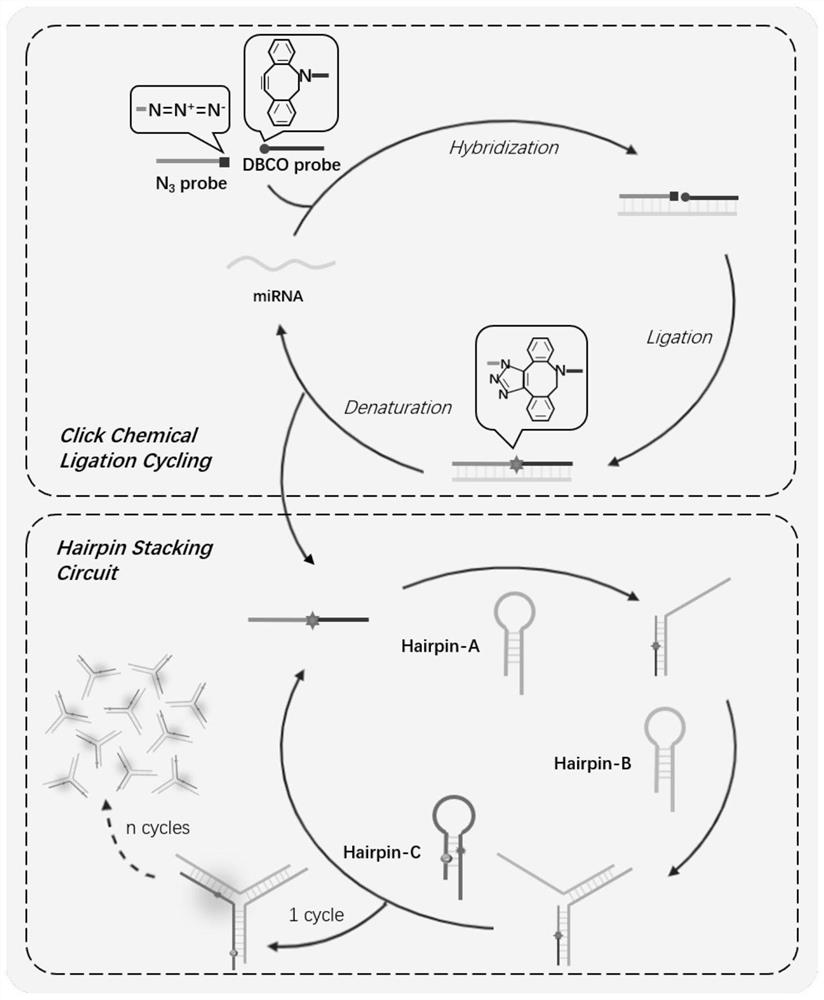
[0044] Example 2. Combining click chemical ligation and hairpin stack assembly to detect microRNA
[0045] figure 1 The implementation process of the present invention is described in detail. When the target exists, two nucleic acid probes (N3 Probe, DBCOProbe) that are respectively complementary to the half sequence of the miRNA molecule Let-7a and are respectively modified with Azide and DBCO groups at the ends can Hybridize with the target so that the click chemical group is close to an efficient click chemical ligation reaction. When the temperature is raised to 95 degrees Celsius, the hybridization complex will be denatured and let-7a will be released, so that it can serve as the next reaction after the temperature is lowered to 30 degrees Celsius. template. After multiple cycles of heating and cooling, a small amount of targets can produce a large number of ligated products. Then we designed three exquisite metastable DNA hairpin structures HA, HB, and HC for the ligate...
Embodiment 3
[0048] Embodiment 3, sensitivity verification
[0049] The sensitivity of the detection method was verified, and the experimental results were as follows: image 3 As shown, in this detection strategy, when the target concentration is in the range of 20pM to 2nM, the fluorescence signal response shows a good linear relationship, and the linear fitting equation is △F=0.3336C+13.544, R 2 =0.9981, the lower detection limit reached 8.63pM. Further, in order to verify that the double-cycle signal amplification we proposed indeed has the effect of double signal amplification, we verified the sensitivity of a single hairpin stack assembly to detect microRNA, using "miR-analog (SEQ ID NO.14)" As a trigger target, results such as Figure 4 As shown, the linear range is 2nM-100nM, and the lower limit of detection is only 1.4nM, which further proves that in the present invention, the addition of the cyclic click chemistry ligation reaction amplifies the signal by more than 100 times an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com