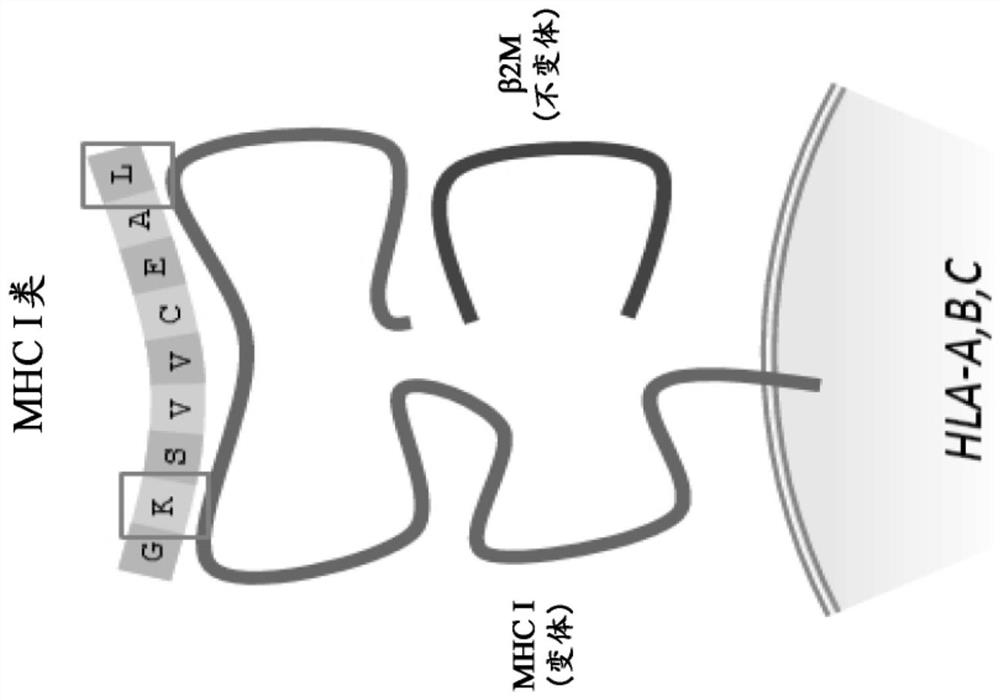
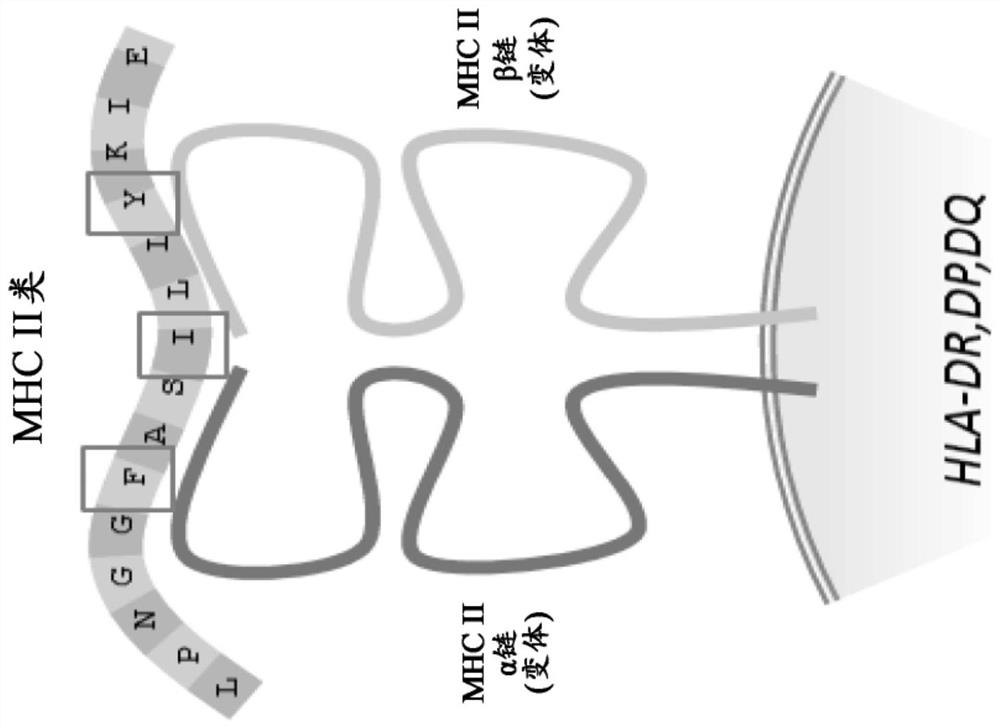
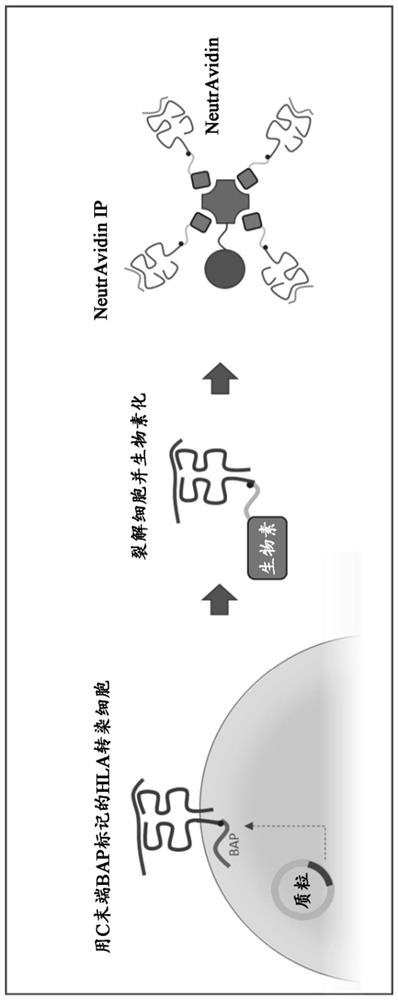
Method and systems for prediction of HLA class ii-specific epitopes and characterization of cd4+ t cells
A cell and prediction model technology, applied in biological systems, chemical instruments and methods, used to analyze two-dimensional or three-dimensional molecular structures, etc., can solve problems that have not yet been fully characterized
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0622] Example 1. HLA II class binding predictor performance
[0623] In this embodiment, the proven data set containing the observed mass spectromethope peptide and the bait peptide is used to analyze the combination predictor neonmhc2 (neon) and NETMHCIPAN. Performance Figure 4 ). For the Neon binding predictor, a separate model is constructed for each MHC II allele displayed. The height of the strip shows a positive predictive value (PPV). When the allele is predicted, the allele is sorted by the performance of the model. Compared to NetMHCIPAN, Neon binding predictors show higher PPV in all alleles.
[0624] In this embodiment, the effect of SPI threshold on the verification of the combined predictor is also tested. Figure 5 ). The HLA Group II class combines the performance of the predictor when training / verification is performed on a group peptide having different score intensity (SPI) cutoffs. Different SPI cutoff conditions: Use the observed MS hit peptide of 70SPI to tr...
Embodiment 2
[0629]Example 2. Neural Network Architecture
[0630] In this embodiment, a neural network is used to obtain a training algorithm ( Figure 9 ). The input peptide is represented as a 20 polymer, and a shorter peptide is filled with "missing" character. Each peptide has 31 dimensional embedded, so input to the neural network is a 20x31 matrix. Before being treated by the neural network, the 20x31 matrix is normalized according to the feature value average and the standard deviation in the training set. The first convolution layer has 9 amino acids and 50 filters (also referred to as channels), with RELU activation functions. Next, it is normalized, then the space is discarded, and the discard rate is 20%. Next, another convolution layer has 3 cores and 20 filters, and the RELU activation function, and then the batch normalization and spatial discard, the discard rate is 20%. The global maximum pool is then applied, and the maximum activated neurons are obtained in each filter in 2...
Embodiment 3
[0631] Example 3. Scalable scheme for the analysis of mono-alleles MHC II ligand
[0632] The knowledge of the MHC II-based binding motif can be calculated based on two in vitro binding tests, and the EC50 is calculated using a cell MHC, and another use of purified MHC calculated IC50. The leading HLA II prediction algorithm NETMHCIPAN is specially trained on this data.
[0633] Limited number of human HLA II alleles currently get more than 200 examples of confirmed combined peptide (affinity Figure 12e ) They are almost a 15 polymer. These experiments cover only the most common colors HLA-DR alleles, limited to non-Caucasic group specific alleles (for example, HLA-DRB1 * 15: 02) coverage, and there is almost no cover of common HLA-DP And HLA-DQ alleles. The current HLA II prediction performance, even in the common high Caucasian allele, it is clearly lagging behind the accuracy of the MHC I class; the ROC curve is only slightly better than the random curve.
[0634] Considering t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com