Method for detecting and typing repeat number of short tandem repeat sequence
A typing method and a technology of repeated numbers, applied in the field of bioinformatics analysis, can solve problems such as a lot of difference (sometimes the ratio of the two exceeds 1:10, inaccurate typing of STR typing detection, multi-STR typing, etc. Achieve high accuracy, strong objectivity, and high data utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Embodiment 1 Method system construction of the present invention
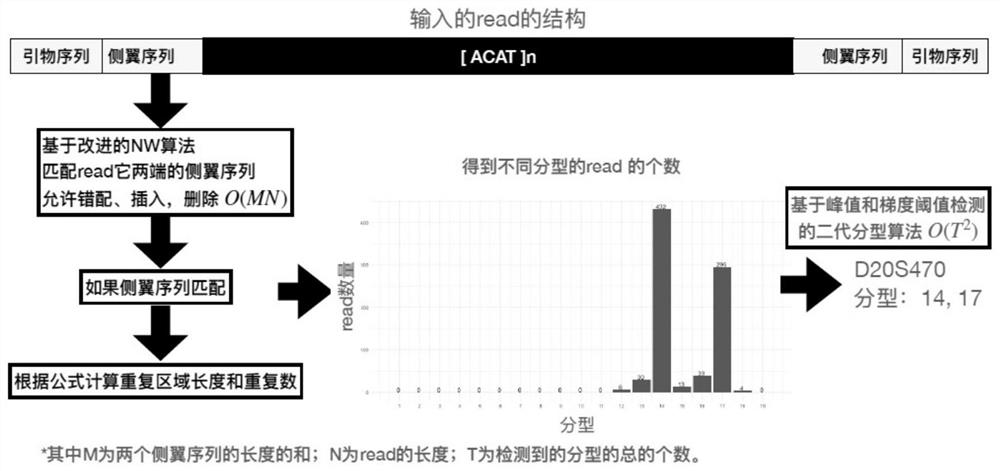
[0090] The process flow of the overall analysis of the next-generation sequencing STR of the present invention is as follows: figure 1 , in the process, first split the off-machine data BCL file into fastq, and then perform read comparison to obtain the STR site to which each read belongs. For all the reads aligned to a certain site, use flanking sequence matching, and if matched, use the repeat number calculation formula to calculate the number of repeats contained in the repeat region. Then count the number of reads of each type in each site, and finally use the typing algorithm to obtain the correct typing. The key steps are the detection of repeated regions and the determination of typing, as follows.
[0091] 1. Construction of search / alignment method for STR flanking sequences
[0092] When using flanking sequences to match reads, the flanking sequences may contain mismatches, insertions and del...
Embodiment 2
[0120] Example 2 Sample Detection Test
[0121] After constructing the algorithm and obtaining the optimal parameter system, the present invention respectively uses standard samples and real samples to test the method of the present invention. Specifically, standard positive samples 9948 and 9947 cell lines, and more than 70 groups of real samples were used for testing. The positive control for real sample testing is the result of capillary electrophoresis (CE). The specific steps include firstly the second-generation library construction and high-throughput sequencing of STR sites; data splitting and sequence comparison after off-machine; finally, the method proposed by the present invention is used to detect the number of repetitions and type STRs, The old method (direct typing through ACR value) was used as a control.
[0122] For the test results of standard samples:
[0123] Using the new method to detect STR typing at 66 loci of 9948 cell lines, the concordance rate b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com