Molecular monitoring method for long-spiny starfish larvae in coral reef area
A technology for coral reefs and thorn sea stars, which is applied in the field of quantitative monitoring of biomolecular markers, and can solve problems such as monitoring of larvae of thorn sea stars that have not been seen.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] (1) Preparation of standard reference DNA template of long spiny starfish larvae
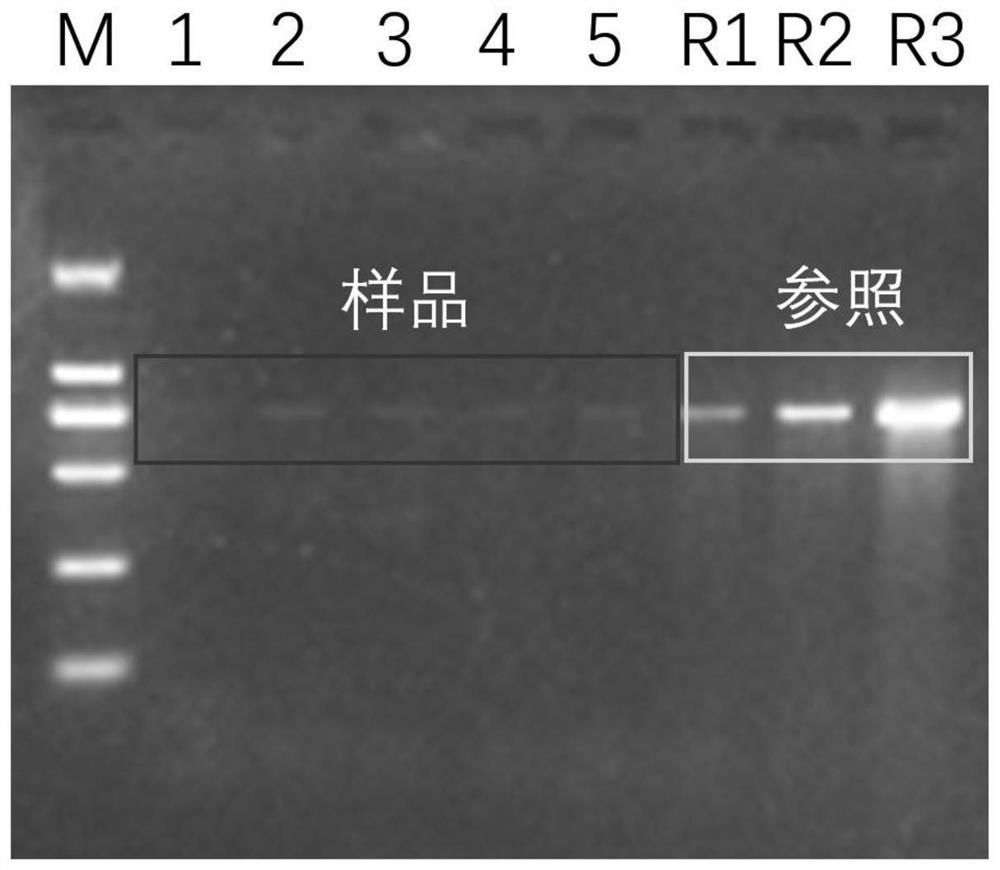
[0023] Quantitatively take the larvae of the long spiny starfish and the brachybrachia larvae for suspension culture, collect about 10,000 samples of the brachycarpus larvae, and filter them with a 100 μm diameter pore / 50 mm diameter nylon mesh filter membrane (Millipore, Germany). Take out the filter membrane with sterile tweezers, and use The filter membrane was cut into pieces with sterile scissors, and together with the filter membrane, the DNA of the biological sample on the filter membrane was extracted using a bacterial genome DNA extraction kit (TaKaRa, China) to obtain the DNA of the larval sample. Take 1 μL of larval sample DNA (approximately 100 larval DNA copies / μL) for serial dilution (5-fold, 10-fold, 20-fold dilution) as a positive control template, and each μL positive control template represents 20, 10, and 5 larvae, respectively DNA copy.
[0024] (2) Filtration and col...
Embodiment 2
[0036] (1) Preparation of standard reference larval DNA template
[0037] Quantitatively take the larvae of the long spiny starfish and the brachybrachia larvae for suspension culture, collect about 10,000 samples of the brachycarpus larvae, and filter them with a 100 μm diameter pore / 50 mm diameter nylon mesh filter membrane (Millipore, Germany). Take out the filter membrane with sterile tweezers, and use The filter membrane was cut into pieces with sterile scissors, and together with the filter membrane, the DNA of the biological sample on the filter membrane was extracted using a bacterial genome DNA extraction kit (TaKaRa, China) to obtain the DNA of the larval sample. Take 1 μL of larval sample DNA (approximately 100 larval DNA copies / μL) for serial dilution (5-fold, 10-fold, 20-fold dilution) as a positive control template, and each μL positive control template represents 20, 10, and 5 larvae, respectively DNA copy.
[0038] (2) Filtration and collection of zooplankton ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com