A new method for preparing short tandem repeat allelic ladders
A technology of short tandem repeats and alleles, which is applied in the field of preparation of allele ladders, can solve the problems of difficult collection of alleles, poor balance of different alleles, and high environmental requirements of the production workshop, so as to avoid template quality differences, Reduce the number of PCR reactions and save manpower and material resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Taking the vWA locus as an example to prepare an allelic ladder
[0055] 1. Experimental method
[0056] 1. Design and synthesize PCR amplification primers according to the reference sequence of the vWA locus, the reference sequence is shown in SEQ ID NO:1.
[0057] The primers include a forward primer F and a reverse primer R; wherein, the 5' end of the forward primer F is modified with a VIC fluorescent dye. The primer sequences are as follows:
[0058] Forward primer F: 5'-TGACTTGGCTGAGATGTG-3';
[0059] Reverse primer R: 5'-GGTTAGATAGAGATAGGACAGA-3'.
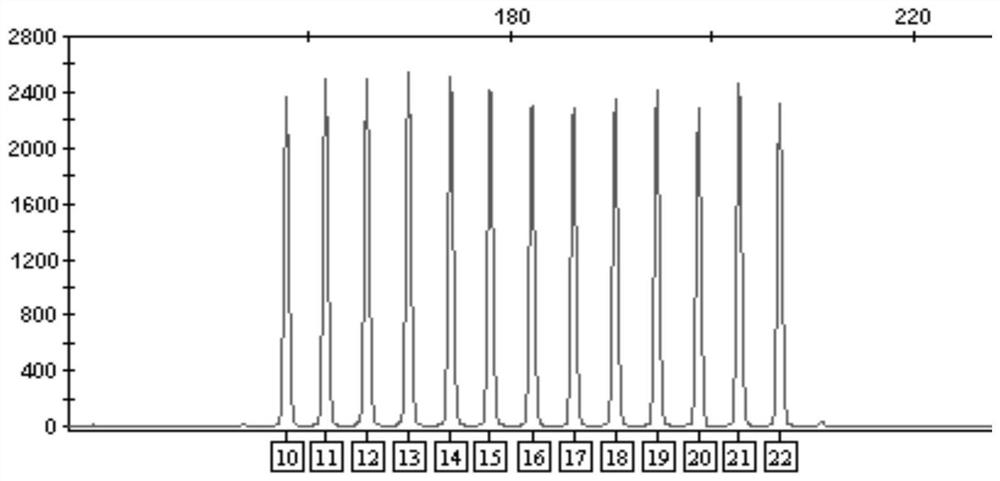
[0060] 2. Design the allelic sequence according to the reference sequence of the vWA locus, and entrust a third-party company to synthesize the plasmid according to the designed sequence. A total of 13 plasmids were synthesized according to the vWA locus, and the core unit [TCTA] repeat numbers were: 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22.
[0061] 3. After the plasmid was synthesized, it wa...
Embodiment 2
[0085] Example 2 Taking the D7S820 locus as an example to prepare an allelic ladder
[0086] 1. Experimental method
[0087] 1. Design and synthesize PCR amplification primers according to the reference sequence of the D7S820 locus, the reference sequence is shown in SEQ ID NO:2.
[0088] Primers include forward primer F and reverse primer R; where the 5' end of forward primer F is modified with FAM. The primer sequences are as follows:
[0089] Forward primer F: 5'-CAGGCTGACTATGGAGTTAT-3';
[0090] Reverse primer R: 5'-ATCCTCATTGACAGAATTGC-3'.
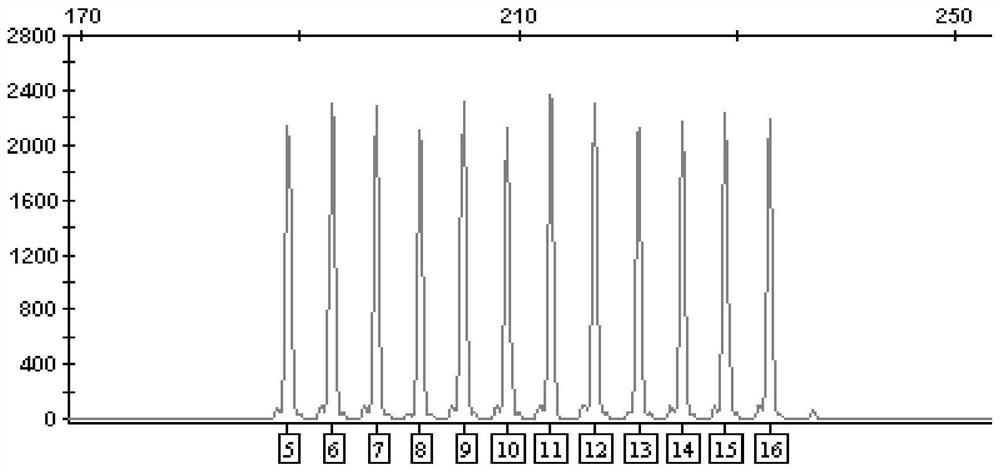
[0091] 2. Design the allelic sequence according to the reference sequence of the D7S820 locus, and entrust a third-party company to synthesize the plasmid according to the designed sequence. A total of 12 plasmids were synthesized at the D7S820 locus, and the repeat numbers of the core unit [TCTA] were: 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, and 16.
[0092] 3. After the plasmid was synthesized, it was diluted to 100 ng / μL with T...
Embodiment 3
[0107] Example 3 Taking the D8S1179 locus as an example to prepare an allelic ladder
[0108] 1. Experimental method
[0109] 1. Design and synthesize PCR amplification primers according to the reference sequence of the D8S1179 locus, the reference sequence is shown in SEQ ID NO:3.
[0110] The primers include a forward primer F and a reverse primer R; wherein the 5' end of the forward primer F is modified with a PET fluorescent dye. The primer sequences are as follows:
[0111] Forward primer F: 5'-CACGGCCTGGCAACTTATATG-3';
[0112] Reverse primer R: 5'-GGATGTGGAGAAACTGAAACCCT-3'.
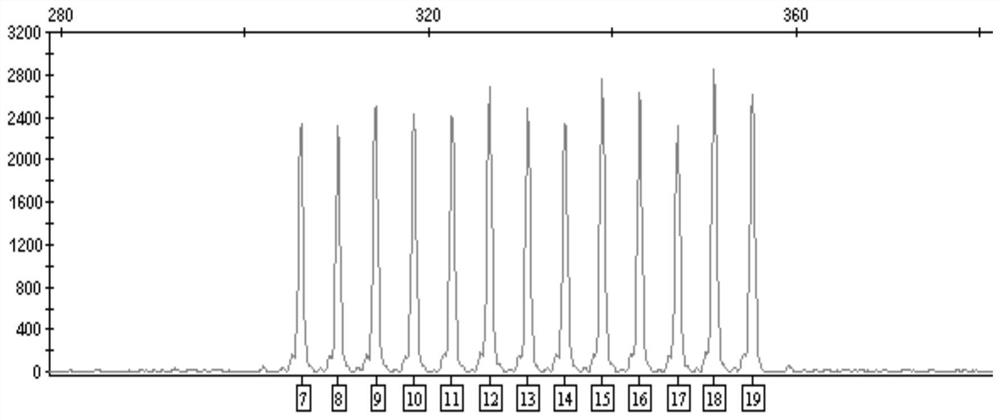
[0113] 2. Design the allelic sequence according to the reference sequence of the D8S1179 locus, and entrust a third-party company to synthesize the plasmid according to the designed sequence. A total of 13 plasmids were synthesized at the D8S1179 locus, and the repeat numbers of the core unit [TCTA] were: 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, and 19.
[0114] 3. After the plasmid is sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com