Kit for rapidly diagnosing peach bacterial shot hole disease
A kit, peach and plum technology, applied in the directions of microorganism-based methods, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of diagnostic errors, cumbersome operation, inapplicability, etc., and meet the requirements of low and high detection equipment. The effect of amplification efficiency and fast extraction speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
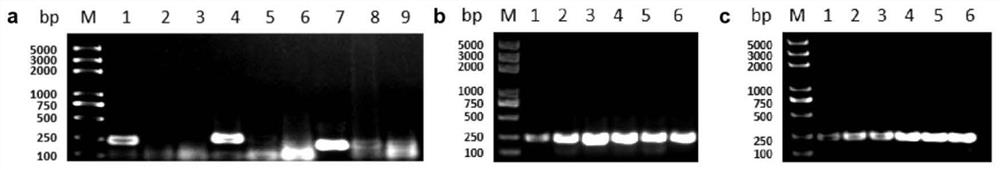
[0049] Example 1 Screening of Xap bacteria RPA detection primers
[0050] When the inventor collected the disease samples of peach bacterial perforation disease from the major peach producing areas in my country, Pantoea sp. bacteria were often isolated from the lesions of peach bacterial perforation disease, and the isolation rate was much greater than that of Xap, as shown in Table 1. They often appear together, and they are morphologically similar, all of which are yellow, raised, smooth-surfaced round colonies, which are difficult to distinguish. However, after a series of identification and verification of Koch's law, it was found that Pantoea sp. was not a pathogenic bacterium, but it caused great trouble to the diagnosis of bacterial perforation disease.
[0051] Table 1 The strains isolated from peach bacterial perforator disease samples
[0052]
[0053]
[0054] The Crispr / Cas12a technique used in the present invention specifically recognizes the PAM sequence (...
Embodiment 2
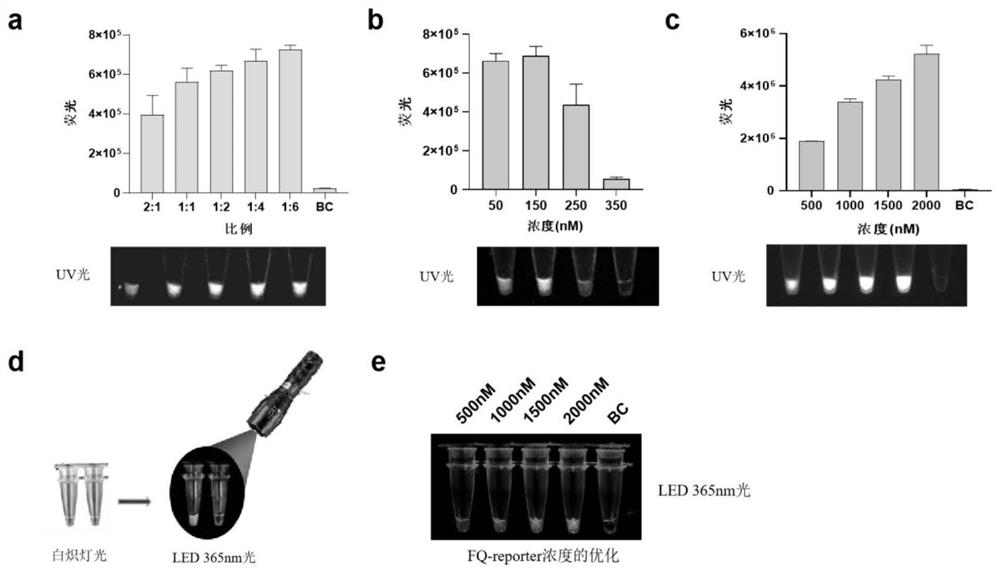
[0060] The optimization of embodiment 2 RPA reaction parameters
[0061] 1. Optimization of the reaction time: the incubation temperature was kept at 37° C., and the reaction time was set to 5 min, 10 min, 15 min, 20 min, 25 min, and 30 min to carry out the RPA reaction respectively (Example 1). The result shows that from 5min to 30min, Xap bacterial strain can be amplified, and the amplification efficiency is gradually enhanced, reaching saturation state in 15min, so 15min is selected as the optimal reaction time (see figure 1 Figure b).
[0062] 2. Optimization of reaction temperature: set 30°C; 33°C; 35°C; 37°C; 39°C; . The results show that the six temperature gradients set can successfully activate the RPA reaction, and the amplification efficiency increases continuously with the increase of temperature, and a high amplification efficiency can be achieved at 37°C, so 37°C is selected as the subsequent reaction temperature (see figure 1 Figure c).
Embodiment 3
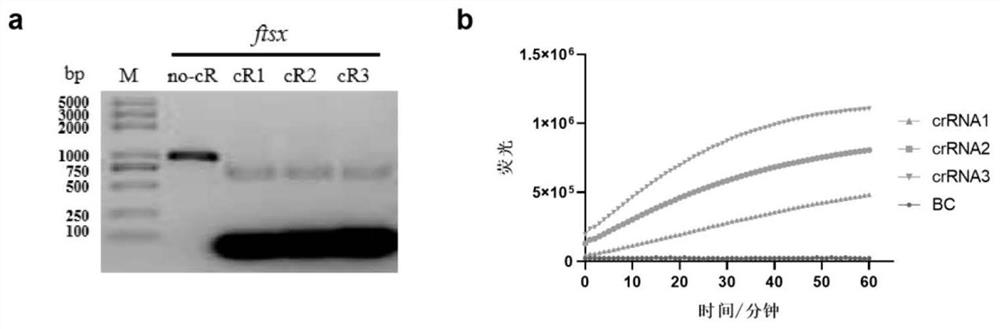
[0063] Example 3 Screening of crRNA for detection of Xap bacteria RPA-Cas12a
[0064] Three crRNAs were designed and synthesized according to the PAM site contained in the target sequence of the optimal RPA1F / 1R primer, and the crRNA guide sequence is shown in Table 3. The in vitro cleavage reaction of dsDNA substrate and the preliminary fluorescence detection reaction of RPA-Cas12 were used to verify the dsDNA and ssDNA cleavage activity of different crRNA-guided Cas12.
[0065] dsDNA substrate in vitro cleavage reaction system:
[0066]1. dsDNA substrate synthesis (ftsx gene): The primer pair XapY17-F and XapY17-R was used. PCR amplification system (total system is 25 microliters): 2×Hieff TM PCR Master Mix (YEASEN, Shanghai) 12.5 microliters, Primer F (upstream primer) 1 microliter, Primer R (downstream primer) 1 microliter, DNA template 1 microliter, dd H 2 O 9.5 μl. PCR reaction program: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 30 s, annealing at ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com