Method for rapidly obtaining fragrant rice material by using CRISPR/Cas9 technology
A technology of cas9-badh2 and fragrant rice, applied in the direction of recombinant DNA technology, botany equipment and methods, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: The process of obtaining genome editing materials
[0042] 1. Design of target joints
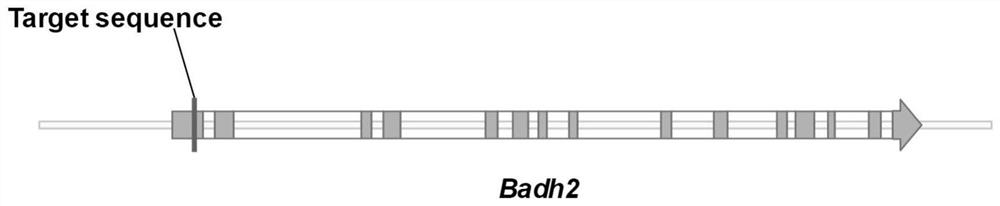
[0043] According to the recognition characteristics of the target site recognized by CRISPR / Cas9, the 26th to 45th bp of the CDS region of the Badh2 gene was selected as the target sequence (the sequence is 5'-AGCTCTTCGTCGCCGGCGAG-3', figure 1 ), the linker primers are as follows:
[0044] Bh2-U3-F: 5'-ggcAGCTCTTCGTCGCCGGCGAG-3';
[0045] Bh2-U3-R: 5'-aaacCTCGCCGGCGACGAAGAGC-3';
[0046] The target sequence is driven by the OsU3 gene promoter.
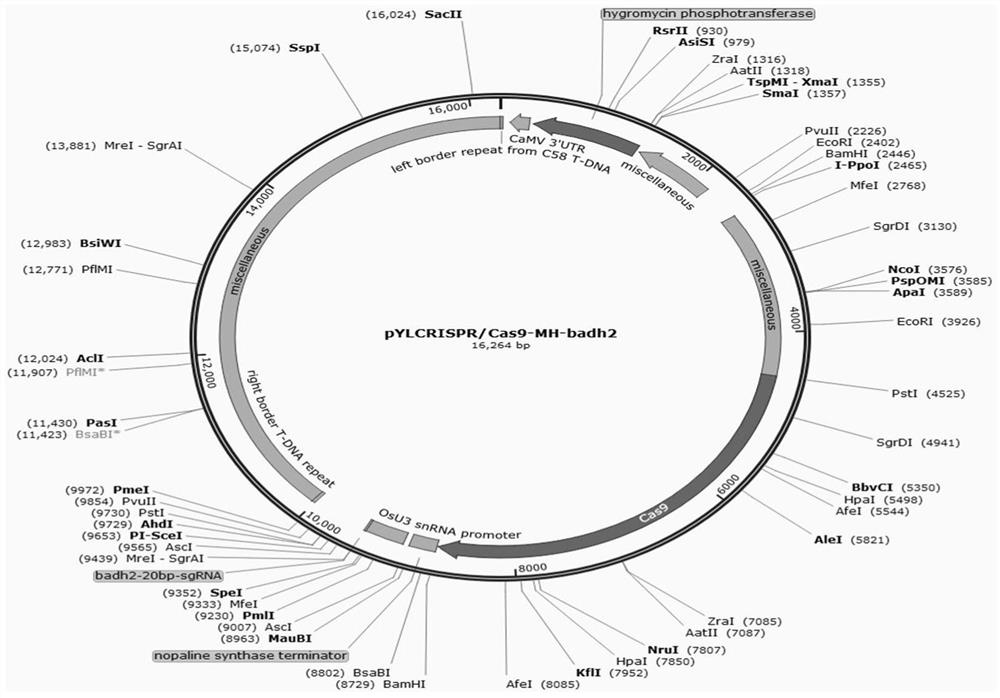
[0047] 2. Construction of pYLCRISPR / Cas9-Badh2 gene editing vector
[0048] The construction of the gene editing vector refers to the method reported by Mao et al. (Mao Y, et al., Mol Plant, 2013, 6(6): 2008-2011.), and proceeds as follows:
[0049] (1) Target linker preparation
[0050] Dissolve linker primers (Bh2-U3-F: 5'-ggcAGCTCTTCGTCGCCGGCGAG-3'; Bh2-U3-R: 5'-aaacCTCGCCGGCGACGAAGAGC-3') with 1x TE (pH8.0) into 100 μM st...
Embodiment 2
[0079] Example 2: Obtaining of No T-DNA Plants and Analysis of Badh2 Gene Knockout
[0080] Grow T in the incubator 0Generation of genome edited plants, self-fertilization, and harvesting of seeds. After the seeds break dormancy, soak the seeds, sow them in seedling trays, and place them in the cultivation room. When the seedlings grow to two leaves and one heart, the genomic DNA of the plants is extracted by referring to the CTAB method of Murray et al. (Murray M G, et al. ., Nucleic Acids Research, 1980, 8(19): 4321-4326). Hpt gene primers encoding hygromycin (hyg283-F: 5'-TCCGGAAGTGCTTGACATT-3', hyg283-R: 5'-GTCGTCCATCACAGTTTGC-3'), Cas9 gene primers encoding Cas9 nuclease (Cas9T-F: 5' -AGCGGCAAGACTATCCTCGACT-3', Cas9T-R: 5'-TCAATCCTCTTCATGCGCTCCC-3') and sgRNA-encoding gene primers (pYLU3-F: 5'-CGTCTTCTACTGGTGCTAC-3', pYLsgRNA-R: 5'-AGTTGATAACGGACTAGCCTT-3') for detection .
[0081] The PCR reaction system consists of 2.6 μl of sterile water, 2.0 μl of primers (1.0 μl ...
Embodiment 3
[0087] Embodiment 3 fragrance phenotype identification
[0088] Theoretically, by knocking out the Badh2 gene, 2-acetyl-1-pyrroline (2-AP) can be accumulated, making rice fragrant. The present invention uses the chewing method to identify the phenotypes of genome edited plants. Dry the rice seeds, peel off the chaff, put the brown rice in your mouth, crush it with your teeth, and perceive the aroma of rice according to the sense of taste and smell. After each sample is identified, it is necessary to rinse the mouth with water to improve the accuracy of identification. After identification, the rice of BY1~BY22 strains all had fragrance.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com