Marker for evaluating hepatic lineage cell maturity, biomics kit and construction method
A lineage cell and cell maturation technology, applied in the field of markers for evaluating the maturity of hepatic lineage cells, can solve the problems of limited stem cell proliferation ability, low functional hepatocytes, and high labor costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
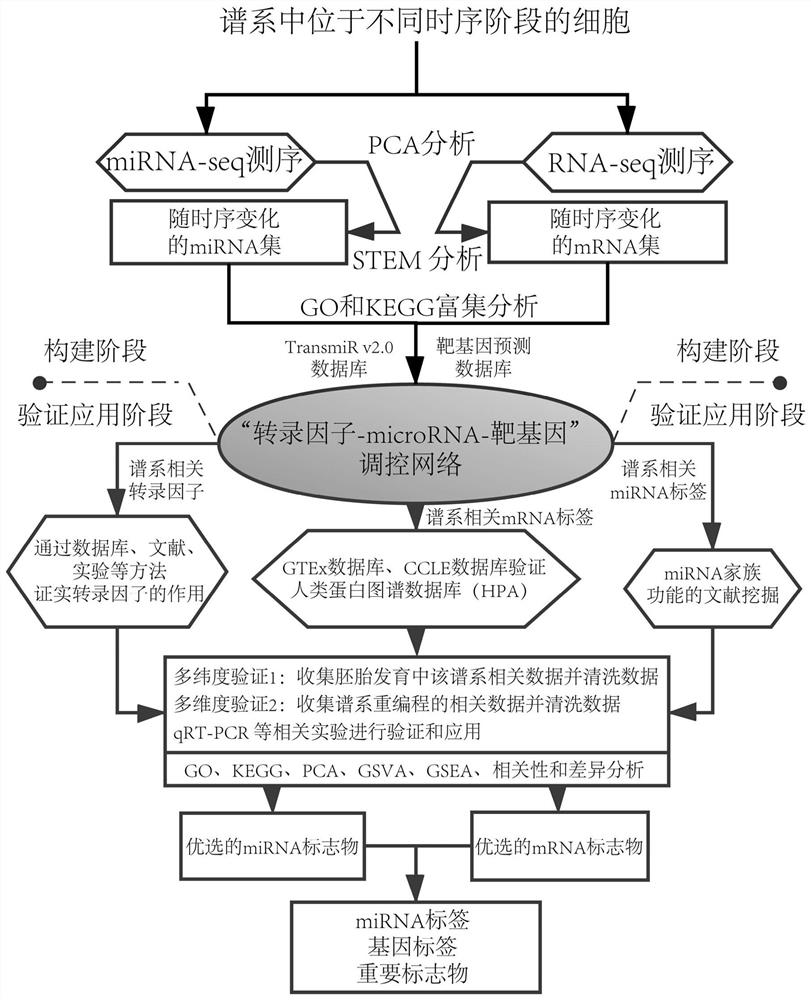
[0157] Example 1. Construction of cell lineage-related "transcription factor-microRNA-target gene" regulatory network
[0158] The method for constructing the regulatory network of "transcription factor-microRNA-target gene" in the process of cell development and maturation lineage mainly includes the following steps ( figure 1 ):
[0159] 1. Obtain cells at different stages of cell lineage; perform microRNA sequencing and RNA-seq sequencing (GSE73114 database in 15 public databases in the aforementioned "1. Preparation of chips and RNA-seq sequencing data of cells at different time series stages in the same lineage" and GSE114974 database after routine processing of sequencing data);
[0160] 2. Through principal component analysis (PCA analysis), it is confirmed that the development of cells does have temporal characteristics;
[0161] 3. Obtain different expression patterns in different cell states through short-time sequence expression analysis (STEM analysis), and obtai...
Embodiment 2
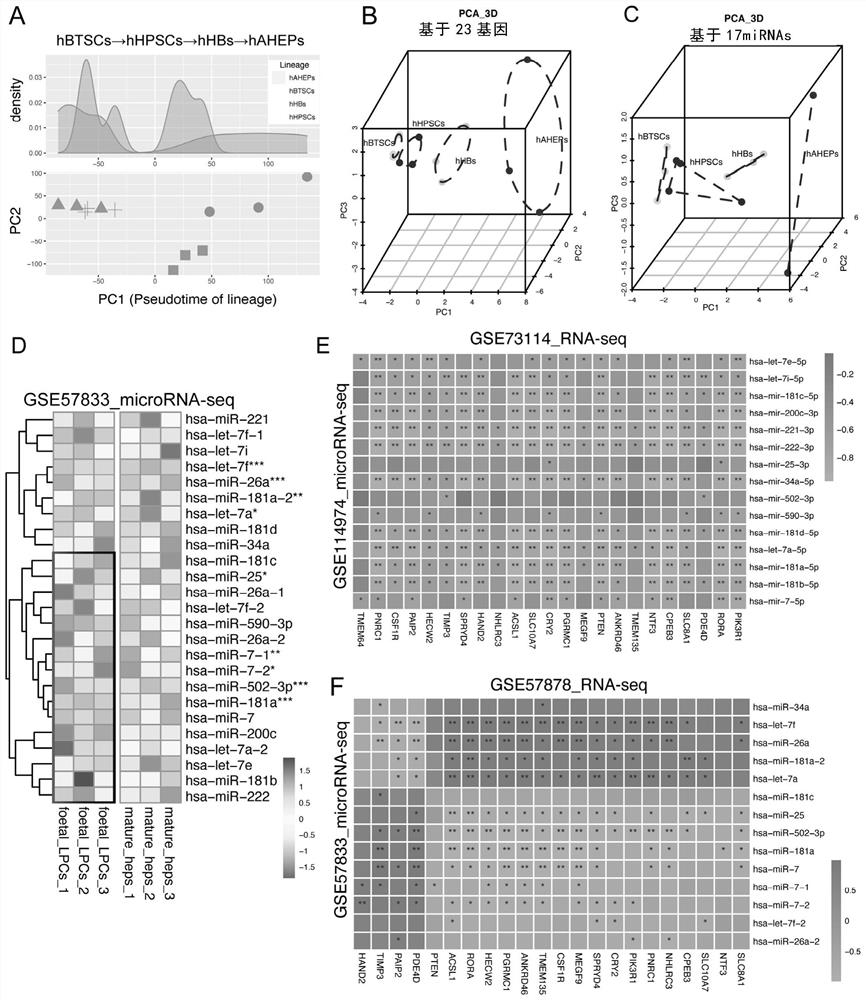
[0176] Example 2. Most of the 17 microRNAs and 23 genes are negatively correlated, and verify the accuracy of the "17microRNA" label
[0177] The inventors analyzed that most of the 17 microRNAs and 23 genes have negative correlations.
[0178] according to image 3 As a result of the PCA analysis in A, it can be seen that the four cell types (biliary tree stem cells (hBTSCs), hepatic stem cells (hHPSCs), hepatic progenitor cells (hHBs) and mature hepatocytes (hAHEPs)) have a very clear timing, so This data is applicable to the "transcription factor-microRNA-target gene" network construction method of the present invention.
[0179] After constructing a network on the public data, the inventors obtained 17 microRNAs and 23 genes, which not only have lineage timing, but also have mutual regulatory relationships. According to the results of 3D_PCA analysis, it can be seen that the differences in timing and maturity of these four cell types can be found only by gene tags or mic...
Embodiment 3
[0182] Example 3. Embryo development-related database verification of the accuracy of the "23 genes" label
[0183] To analyze 23 gene signatures (PIK3R1, PTEN, ACSL1, ANKRD46, CPEB3, CRY2, CSF1R, HAND2, HECW2, MEGF9, NHLRC3, NTF3, PAIP2, PDE4D, PGRMC1, PNRC1, RORA, SLC10A7, SLC8A1, SPRYD4, TIMP3, TMEM135 , TMEM64), the inventors used the following disclosed embryonic development-related databases to obtain cells at different lineage developmental stages, and analyzed the expression of the 23 gene signatures.
[0184] GSE90047: Including cells developed from hepatoblast to hepatocytes during embryonic development, embryonic day 10.5 (E10.5) to embryonic day 18.5 (E18.5);
[0185] GSE132034: including mouse liver organs at various developmental stages, embryo E12.5 → eighth week after birth (W8);
[0186] GSE28892: includes three types of cells: adult hepatic progenitor LPCs (Adult LPCs), differentiated mature hepatocytes (Differentiated Heps), and primary hepatocytes (Primary...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com