Method for creating novel high-amylose rice germplasm and application of novel high-amylose rice germplasm
A high-amylose, new germplasm technology, applied in the field of gene editing, can solve problems such as incomplete clarity, and achieve the effect of low cost and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
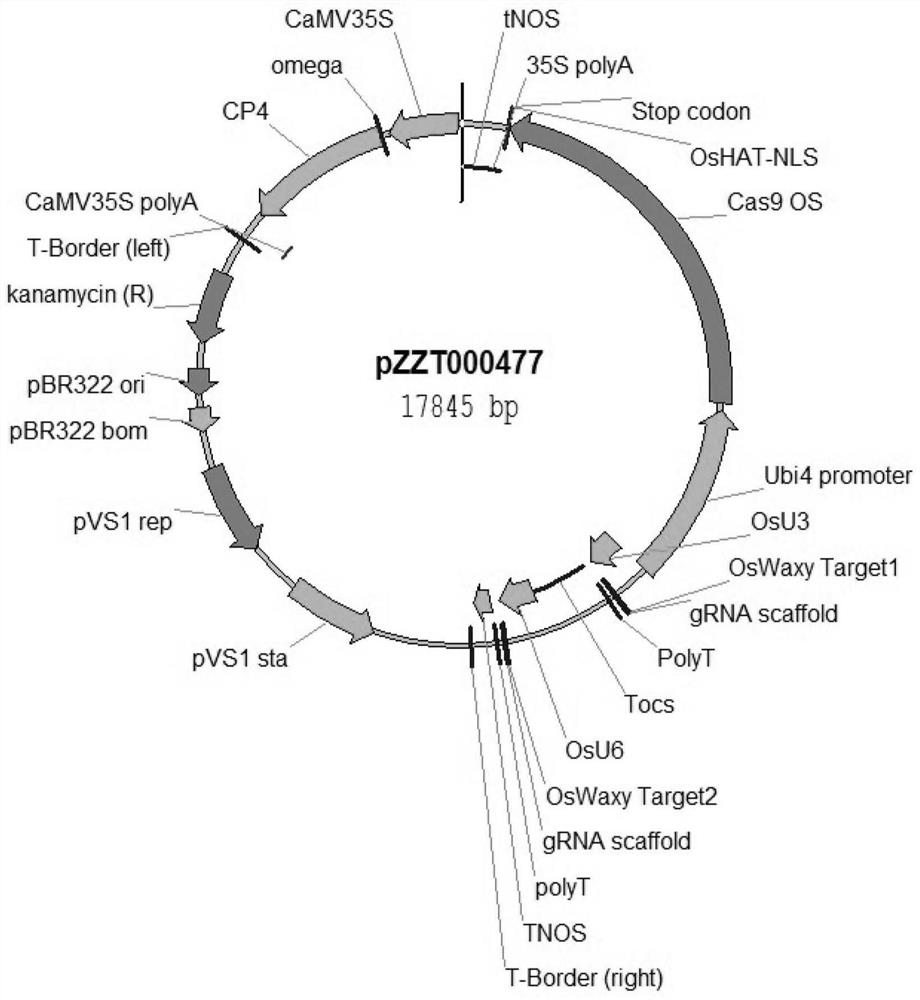
[0076] Example 1 Construction of CRISPR / Cas9 editing vector
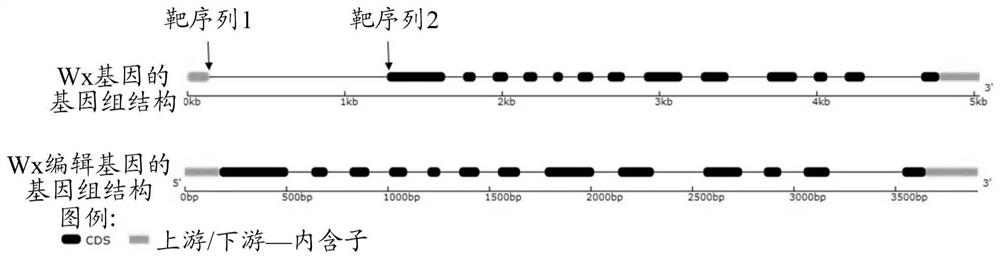
[0077] Sequence analysis and gene editing target design of rice OsWaxy gene
[0078] The sequence of the rice OsWaxy gene is shown in SEQ ID NO:1. Sequence analysis showed that the gene contained 14 exons and 13 introns, of which the first intron was located in the 5'UTR, located at bases 139-1267 in SEQ ID NO:1. The target sequence 1 and target sequence 2 set in this example are both located in the first intron of the OsWaxy gene in the indica rice X32 and Kongyu 131 materials, wherein the target sequence 1 is at the 174-192 position of SEQ ID NO:1 base (i.e. TCTTATTCAGATCGATCAC (SEQ ID NO: 3)), the target sequence 2 is at the 1217-1236th base of SEQ ID NO.1 (GTTCAAATTCTTTTAGGCTC (SEQ ID NO: 4)), such as figure 2 .
[0079] Construction of CRISPR / Cas9 editing vector
[0080] The gene editing technology used in this example is the third generation gene editing technology CRISPR / cas9, the vector used is designed...
Embodiment 2
[0081] Example 2 Genetic transformation of indica rice X32 and japonica rice Kongyu 131
[0082] The editing vector constructed in Example 1 was transformed into Agrobacterium strain EHA105 (a gift from Huazhong Agricultural University) and sequenced for identification. Embryogenic calli were induced from the seeds of indica rice X32 and japonica rice Kongyu 131, respectively, and co-incubated with Agrobacterium EHA105 containing the above-mentioned editing vector, and genetic transformation was completed through a series of steps such as screening and regeneration. Agrobacterium-mediated transformation in rice. Nature protocols, 2006, 6:2 796-2802). The specific process includes, after 3 days of co-cultivation of the rice callus and Agrobacterium, the callus was added to the ddH of 250mg / L Carbenicillin 2 Soak in O for 30 minutes, remove excess Agrobacterium, and place on a screening medium containing 250 mg / L Carbenicillin for 3 rounds of screening, then transfer the resist...
Embodiment 3
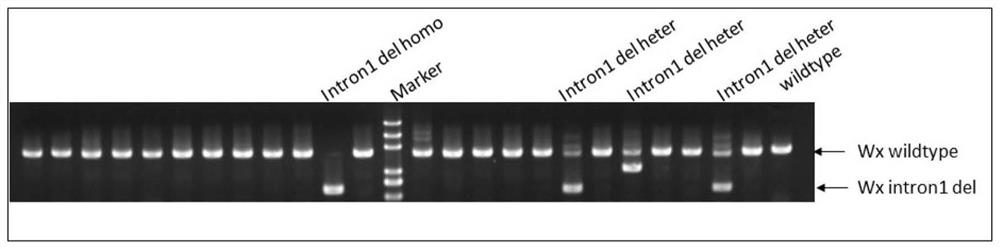
[0083] Example 3 Screening and identification of T0 generation OsWaxy gene-edited rice
[0084]The regenerated strains in Example 2 were sent to the greenhouse for planting, and the leaves of the regenerated T0 generation seedlings were taken, and plant genomic DNA was extracted by the CTAB method as a template. DNA samples were positively detected by fluorescent quantitative PCR method. The fluorescent quantitative PCR method selects the screening marker gene CP4 as the target gene and the rice ACTIN1 gene as the internal reference gene, performs amplification and fluorescence value detection on a fluorescent quantitative PCR instrument, and screens out transgenic positive lines according to the RQ value (CP4 gene RQ value> 0.1, results not shown). The primer sequence used to amplify the screening marker gene CP4 is csp356: CAGCACAGGTTAAGTCTG (SEQ ID NO: 8) and csp357: GTCTGTCTCAACGGTAAG (SEQ ID NO: 9); the primer sequence used to amplify the ACTIN1 gene is csp106: TGCTATGTA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com