Dual-induction mCreER system capable of tracking cell differentiation and development and establishment and application of dual-induction mCreER system
A cell differentiation and traceable technology, applied in the field of cell lineage tracing, can solve the problems of genome damage to cells, reduced reliability of tracing results, leakage, etc., to achieve the effect of improving tissue specificity, reducing off-target phenomena, and reducing cytotoxicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
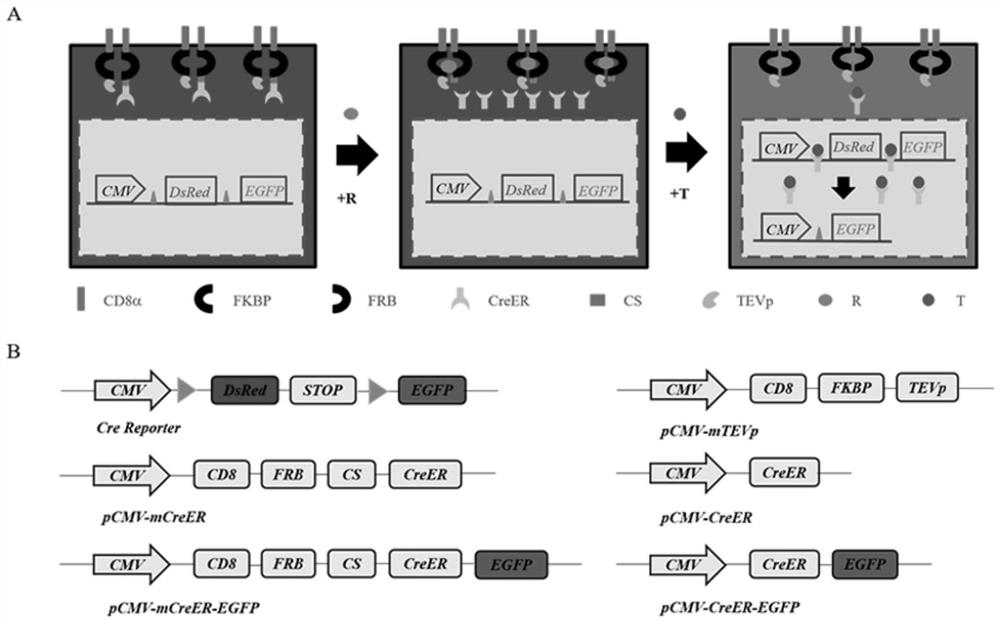
[0037] 1. The establishment principle of a dual-induction mCreER system that can track cell differentiation and development is as follows: figure 1 As shown in A, the mCreER and mTEVp proteins are respectively connected to CD8 located on the cell membrane through the FRB protein and the FKBP protein, and are correspondingly fixed on the cell membrane; when the inducer rapamycin is added, the FKBP protein and the FRB protein Close to each other and combine to form a complex, induce the cleavage of mTEVp protein, release mCreER from the plasma membrane, and enter the nucleus under the action of the inducer tamoxifen to mediate loxP site recombination, realizing the controllability of recombination time. The gene sequence of the mCreER is shown in the sequence listing SEQ ID NO:1; the gene sequence of the mTEVp protein is shown in the sequence listing SEQ ID NO:2. The gene of the mCreER must encode the amino acid sequence shown in SEQ ID NO: 3 in the sequence listing; the gene of...
Embodiment 2
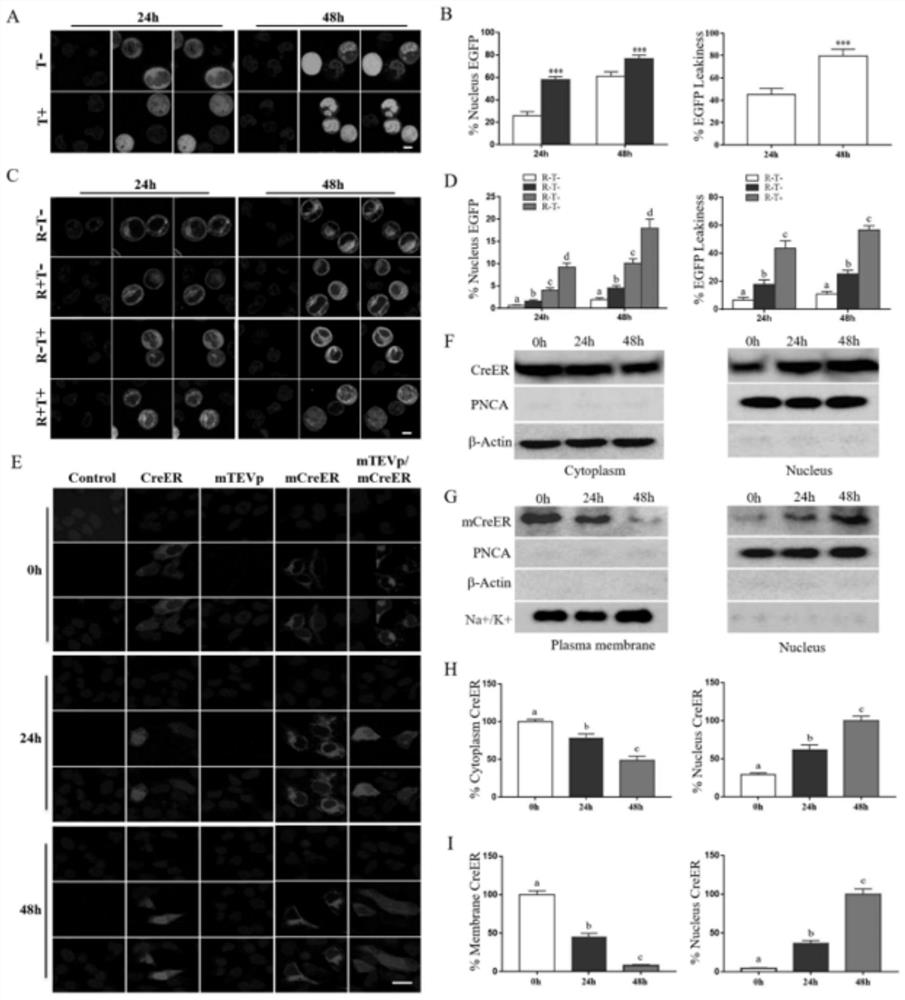
[0055] Intracellular localization verification of traditional CreER system and dual-inducible mCreER system.
[0056] 1. Construction of pCMV-CreER-EGFP plasmid: Use Nhe and EcoRI enzymes to obtain the vector backbone of the pCMV-EGFP-N1 plasmid, digest the CreER fragment obtained by PCR amplification, connect the fragment to the above vector backbone, and perform digestion and sequencing After verification, the recombinant pCMV-CreER-EGFP was obtained.
[0057] F-primer(5'→3')R-primer(5'→3')
[0058] CGGCTAGCCGATGGC GAATTCGAGCTGTGGCAGGGA CAATTTACTGACCGTACA AACCC
[0059] 2. Immunofluorescence staining to verify the location of CreER in the cells mainly includes the following steps: (1) After transfecting the cells of the traditional CreER system and the double-induced mCreER system, the cell culture medium was aspirated, washed once with PBS, and 4% polymer was added. Formaldehyde fixation for 15 minutes. (2) Wash three times with PBS, 3 minutes each time, add 0.2% Trito...
Embodiment 3
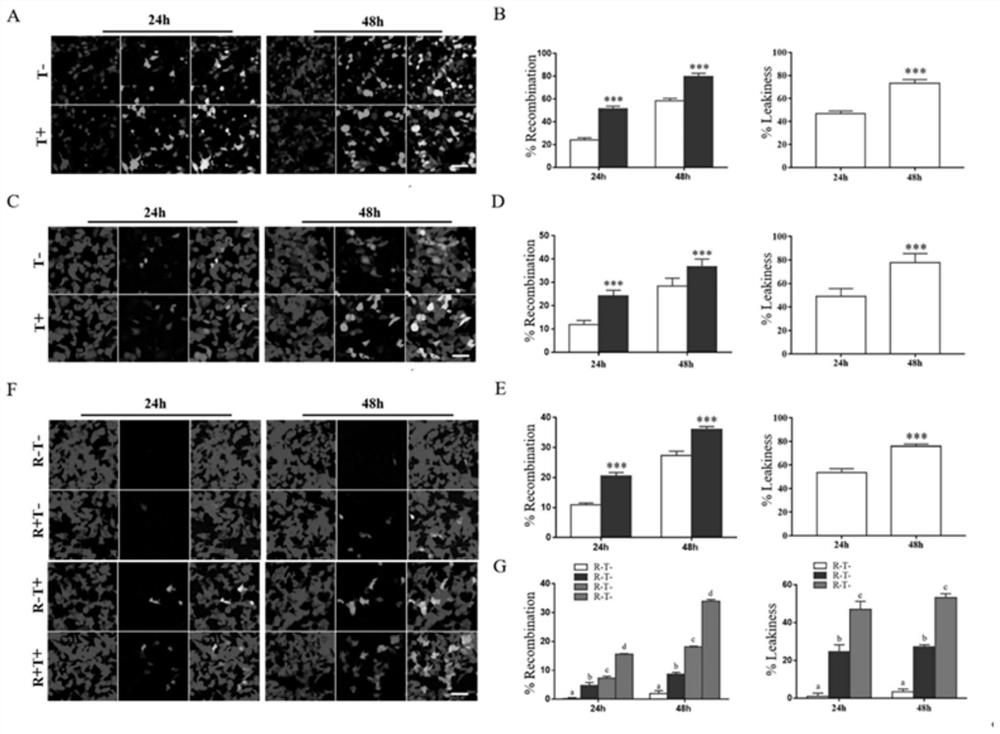
[0063] HSP90 plays an important role in the stringency of the mCreER system
[0064] Ad293 was co-transfected with pCMV-mCreER / pCMV-mTEVp plasmid, the inducer was added 24 hours after transfection, the cells were collected and lysed after 24 hours of induction, and the expression levels of CreER, CD8a and HSP90 were verified by co-immunoprecipitation and WB, indicating that HSP90 has a positive effect on the mCreER system important role. After the HSP90 protein was knocked down by siRNA, the stringency of mCreER system labeling cells was greatly reduced, and serious leakage occurred. It is speculated that HSP90 protein binds to ER, which can seal the CS site and effectively prevent TEVp enzyme from cutting the CS site. When tamoxifen is added, the conformation of ER changes, dissociates from the anchored HSP90, and Under the mediation of nuclear signal, CreER protein enters the nucleus and recombines the Loxp site. The results of studying the mechanism of mCreER system by co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com