Pathogenic microorganism virulence gene association model as well as establishment method and application thereof
A technology of pathogenic microorganisms and virulence genes, applied in the fields of genomics, bioinformatics, instruments, etc., can solve the problems of easy introduction of pollution, limited detection targets of virulence factor detection, and limited results, so as to achieve the detection of virulence. Comprehensive genes, unlimited sample types, and small sampling requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] A method for establishing a pathogenic microorganism virulence gene association model, comprising the following steps:
[0075] 1. Establish a gene database of pathogenic microorganisms.
[0076] Obtaining the reference gene data of pathogenic microorganisms and constructing the pathogenic microorganism gene database can be carried out according to conventional methods. In this example, the pathogenic microorganism genome database constructed by the technical scheme disclosed in the Chinese Invention Patent with the application number of our company is 201910779825.0 for subsequent analysis Comparison.
[0077] It can be understood that the method of the present invention is not limited to specific databases of pathogenic microorganisms, and any open-source and general-purpose databases are acceptable.
[0078] 2. Establish a virulence gene database.
[0079] 2.1 Survey to obtain a list of virulence genes with clear clinical significance.
[0080] Based on the curren...
Embodiment 2
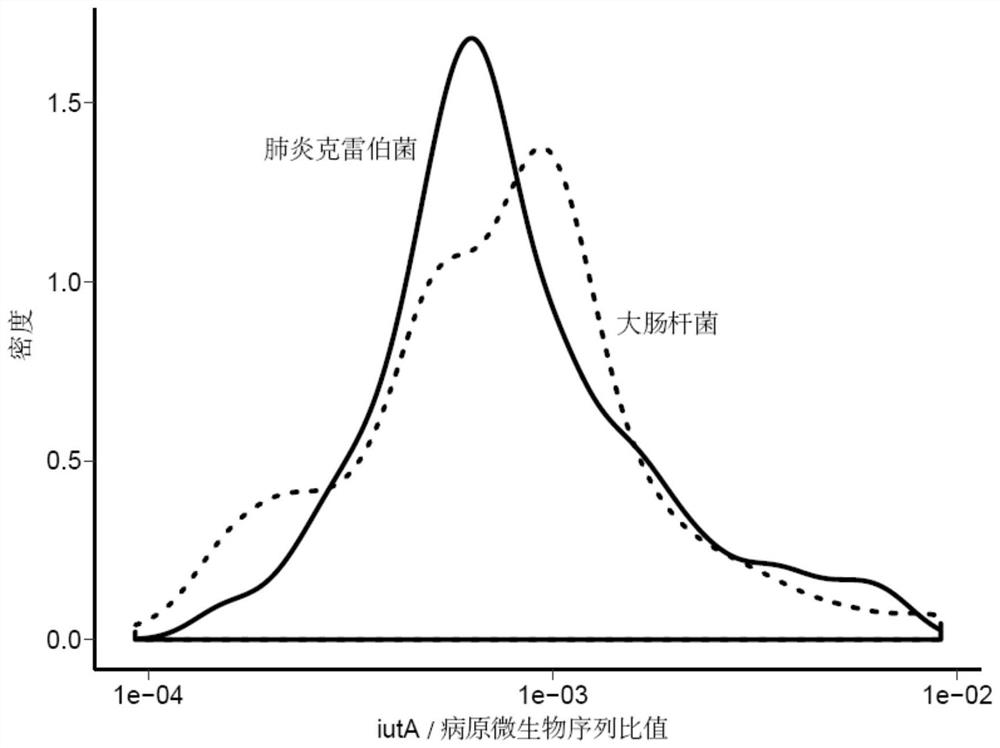
[0113] From the 50,000 clinical samples, 760 samples with Klebsiella pneumoniae detected were selected. First, the sequence ratio of the virulence gene iutA and Klebsiella pneumoniae was calculated, and the log transformation was performed.
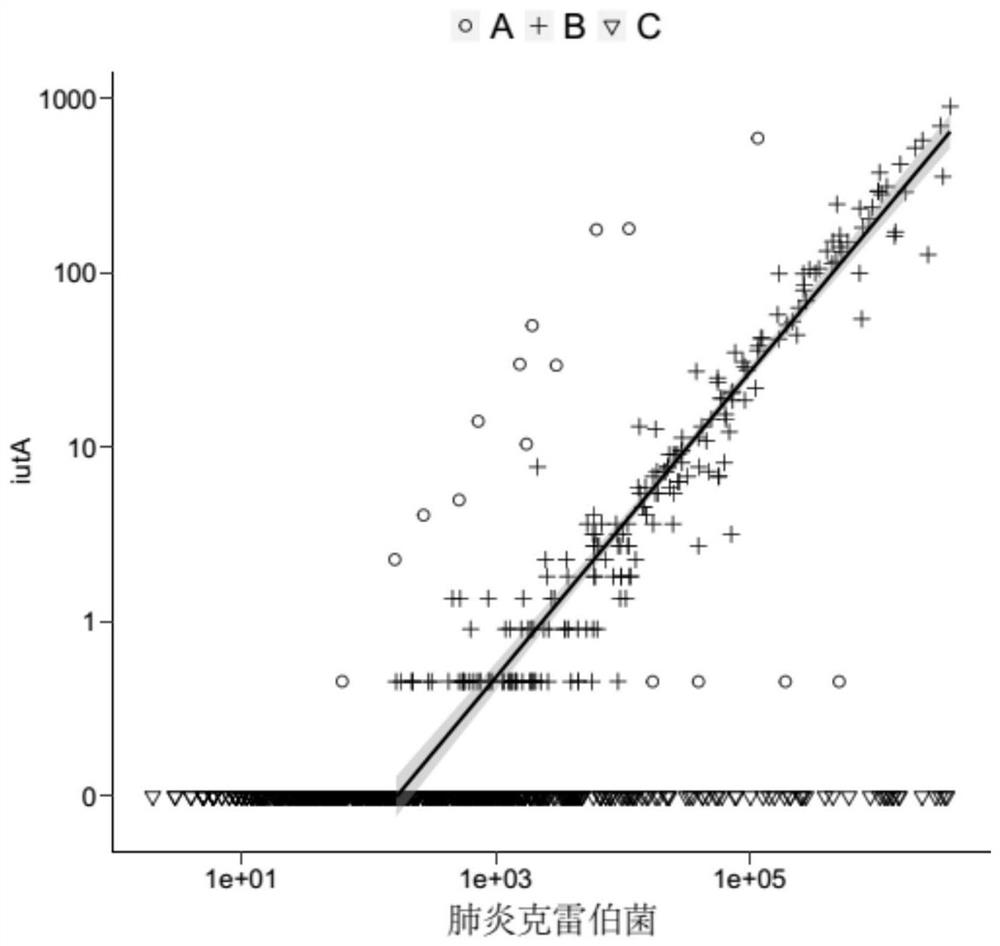
[0114] With the iutA-Klebsiella pneumoniae association model of embodiment 1, the sequence ratio of iutA and Klebsiella pneumoniae Klebsiellapneumoniae is divided into 3 classes (A, B, C), such as image 3 As shown, class B is a sample that satisfies the model (derived from Klebsiella pneumoniae), class A may be a sample from Escherichia coli and Shigella flexneri, and class C is a sample without iutA sequence detection;
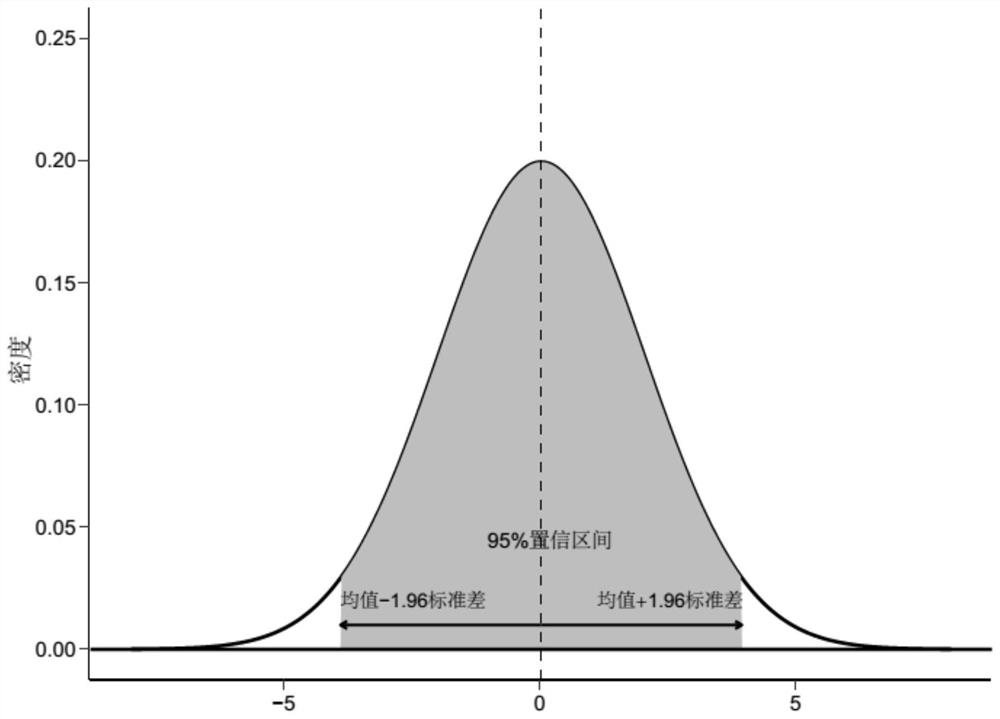
[0115] Figure 4 It is a box plot of the ratio distribution of the iutA gene and Klebsiella pneumoniae, where the upper and lower dashed lines are the 95% confidence interval of the ratio distribution of the sample that satisfies the model, specifically [5.4e-5,3.7e-3]. In our subsequent application, we can judge wheth...
Embodiment 3
[0118] According to the method of the above-mentioned Example 1, based on the data of 50,000 cases of clinical samples, a variety of pathogenic microorganism virulence gene association models were established, and the following 95% confidence intervals for the ratio of each virulence gene to bacteria were obtained for clinical judgment applications.
[0119] Table 2. 95% confidence intervals for the ratio of virulence genes to bacteria
[0120] virulence gene bacteria 95% lower confidence ratio 95% Upper Confidence Ratio wxya Enteropathogenic Escherichia coli 0.0000182 0.001818652 TcdA Clostridium difficile 0.0000233 0.002330865 TcdB Clostridium difficile 0.0000233 0.002330865 eae Enteropathogenic Escherichia coli 0.0000182 0.001818652 eltA Enterotoxigenic Escherichia coli 0.0000182 0.001818652 eltB Enterotoxigenic Escherichia coli 0.0000182 0.001818652 estIa Enterotoxigenic Escherichia coli 0.0000182 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com