Method for amplifying target region of nucleic acid, library building and sequencing methods and kit
A technology for target regions and nucleic acids, applied in the field of gene sequencing, can solve problems such as not reflecting the absolute abundance of species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
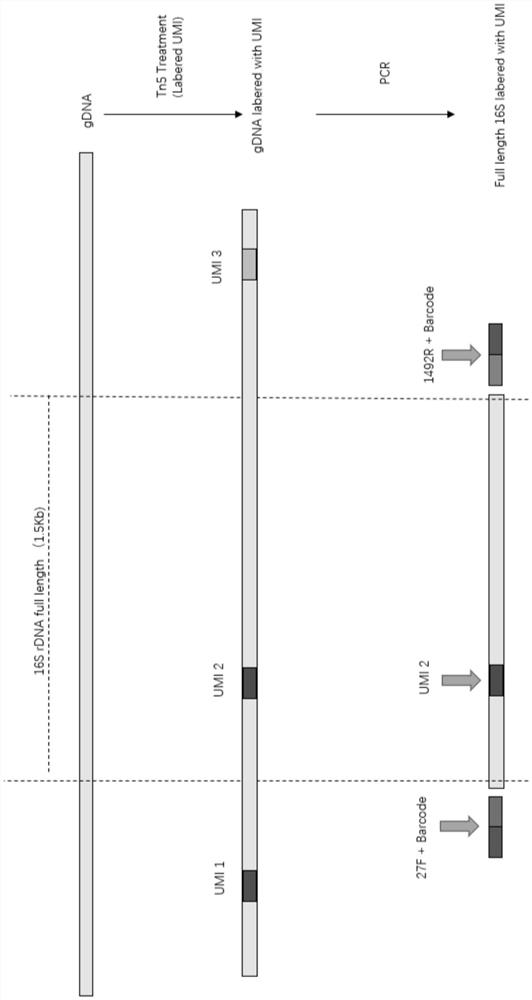
[0052] Embodiment 1 uses Tn5 transposase to carry out the method for library construction and sequencing
[0053] Embodiment 1 provides a kind of method that adopts T5 transposase to carry out library building and sequencing to the 16s DNA of microbial strain, comprises the following steps:
[0054] (1) Sample preparation: use the following four strains (subsequently referred to as A\B\C\D), extract the DNA of each strain, and then mix according to the 16S rDNA copy number of each strain, so that each microliter solution contains 50,000 copies of DNA from each strain.
[0055] Abbreviation of strain Full name of the strain GC content% copy number per μl A Rhodobacter sphaeroides 69.01 50000 B Escherichia coli 50.79 50000 C Acinetobacter baumannii 38.94 50000 D Bacillus cereus 35.58 50000
[0056] (2) Tn5 processing mark:
[0057] A. Synthesize the nucleic acid fragment containing the recognition sequence and UMI sequence (t...
Embodiment 2
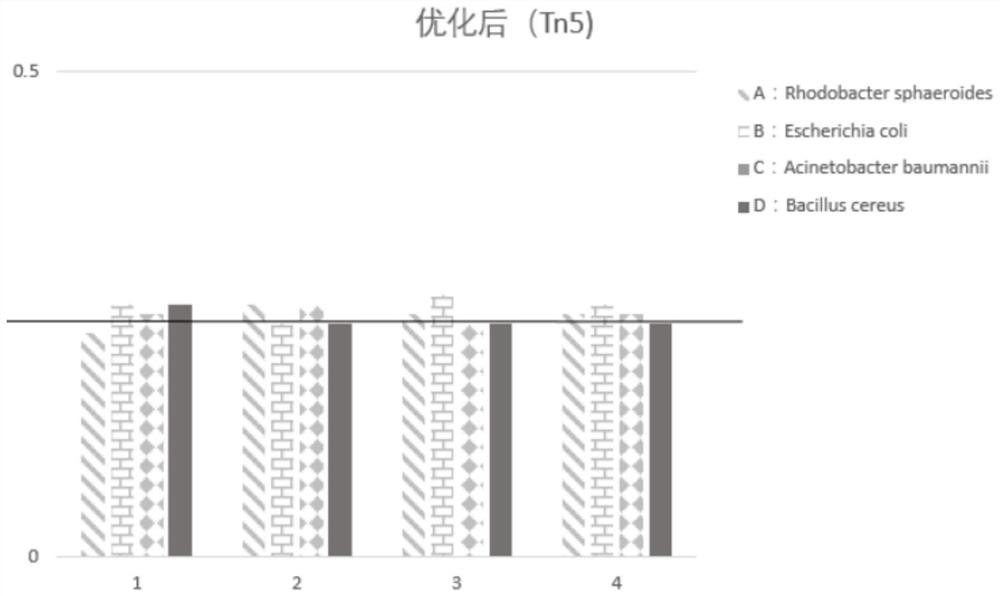
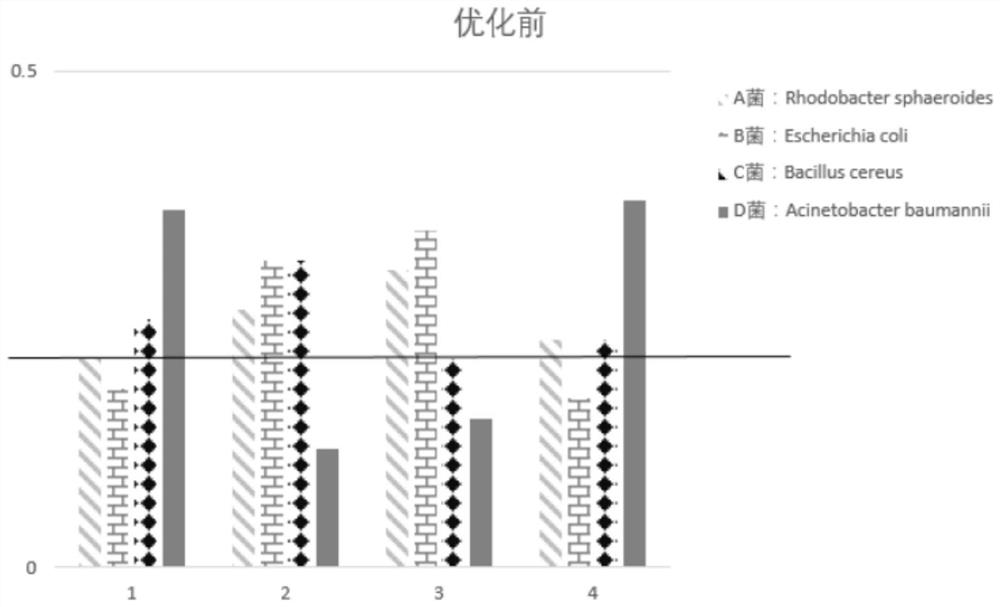
[0107] Embodiment 2Tn5 reaction conditions are explored
[0108] With reference to the method of Example 1, Example 2 explored the reaction conditions of step C when step (2) Tn was used for labeling, respectively at 55 degrees Celsius, 45 degrees Celsius, 37 degrees Celsius, and 20 degrees Celsius. Down reaction 30 minutes, as shown in table 3 below:
[0109] Table 3 Amplification results and data of reaction condition exploration
[0110]
[0111]
[0112] In Table 3, the length before Tn5-UMI treatment represents: the length of the amplified fragment without using Tn5 transposase. There are certain differences in the length of Tn5-UMI before treatment obtained under different reaction conditions. The possible reasons are mainly: when using Agilent2100 for detection, the Agilent2100 instrument itself is relatively sensitive, which brings some differences in the detection results; in addition Genomic gDNA itself is a mixture of multiple bacteria, and there may also be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com