Primer combination, and detection reagent or kit for anisakis
A combination of primers and anisakis technology, applied in the biological field, can solve problems such as contamination of false positive results, complex primer design, environmental pollution, etc., and achieve the effect of portable nucleic acid detection, simple primer design, and large development space
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The identification of embodiment 1 anisakis
[0048] Extraction of parasite genomes (Anisakis, Schizocephalus lacunae, Clonorchis sinensis, Mesorchis orientalis, Echinostomum)
[0049] (1) Take out the parasites from the -80°C refrigerator, freeze and thaw them repeatedly with liquid nitrogen and boiling water for 3-5 times, 30s each time.
[0050] (2) Add 500 μL of lysate to a 1.5 mL centrifuge tube, then add 20 μL of 2% proteinase K, shake and mix.
[0051] (3) Put the centrifuge tube in a 56°C water bath, incubate for 2 hours, and turn the centrifuge tube 3-5 times every hour (the purpose of turning is to fully lyse the tissue).
[0052] (4) Add 5 μL of RNaseA, mix well and let stand for 2 minutes.
[0053] (5) Add an equal volume of Tris-saturated phenol (500 μL), vortex vigorously and mix well, then let stand for 10 min.
[0054] (6) Centrifuge at 12,000rpm at 4°C for 15min. After centrifugation, it is divided into upper, middle and lower layers. The upper layer...
Embodiment 2
[0065] Example 2 Primer Design and RPA Condition Optimization
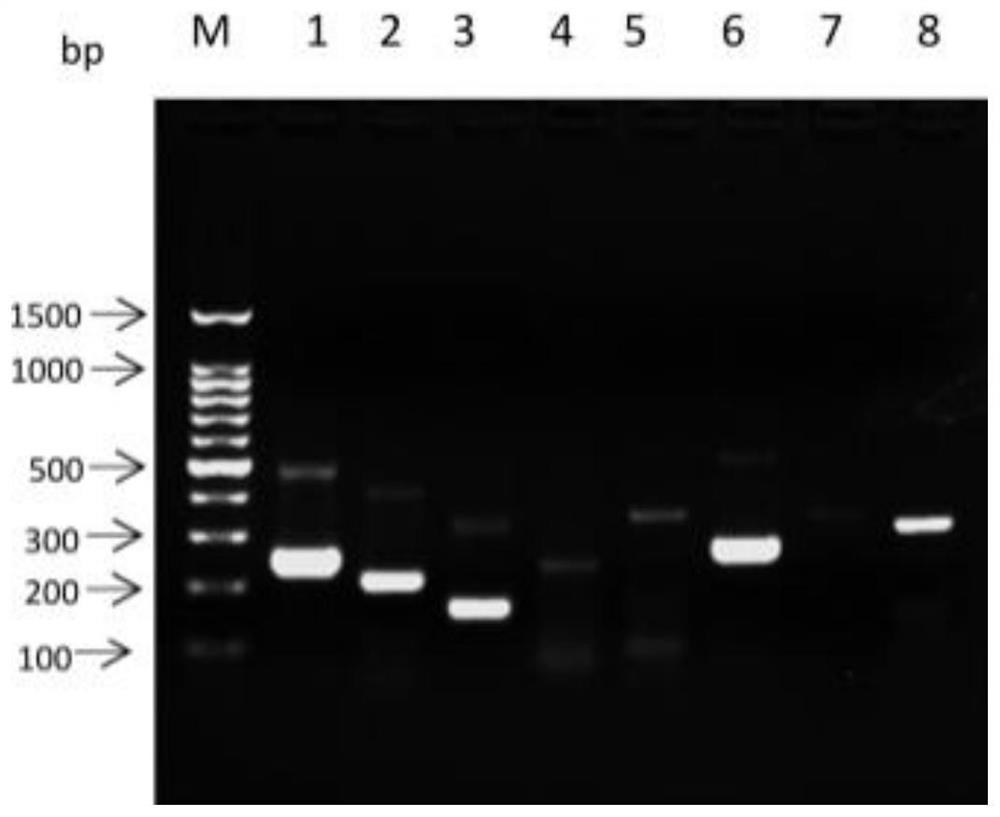
[0066] According to the sequence comparison of the ITS region of the identified insect species by DNAMAN, 8 pairs of RPA amplification primers were designed, namely ITS-1, ITS-2, ITS-3, ITS-4, ITS-5, ITS-6, ITS -7, ITS-8. The sequence is as follows:
[0067] ITS-1: F, 5'-CTAGGTGGCCGCCAAAACCCAAAACACAACC-3';
[0068] R, 5'-ACAGTTCACCGTAATTCGACCCTCAGCCAGACG-3';
[0069] ITS-2: F,5'-CTAGGTGGCCGCCAAAACCCAAAACACAACC-3';
[0070] R, 5'-ACGAACCGAGTGATCCACCGCCAAGATTTG-3';
[0071] ITS-3: F,5'-TGCGATAAATAGTGCGAATTGCAGACACAT-3';
[0072] R,5'-AATTGCTTGCCGAATGCTTCACAGTCCAGAAAAA-3';
[0073] ITS-4: F,5'-GTCAGTTGCGATGAAAGATGCGGAGAAAGTTCC-3';
[0074] R, 5'-TGCTCAATGTGTCTGCAATTCGCACTATTTATCG-3';
[0075] ITS-5: F,5'-TGTTGAACAACGGTGACCAATTTGGCGTCTACG-3';
[0076] R, 5'-AGTGATCCACCGCCAAGATTTGTACATTTTCAACACAT-3';
[0077] ITS-6: F,5'-TGTTGAACAACGGTGACCAATTTGGCGTCTACG-3';
[0078] R, 5'-AGCTGGCTGCGTTTCTTCATCGATCCACGAA-3';...
Embodiment 3
[0091] Embodiment 3 Sensitivity and specificity detection
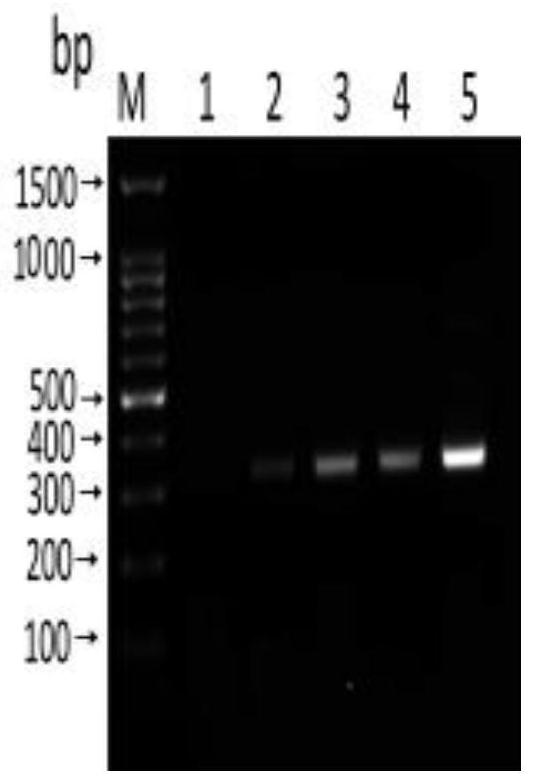
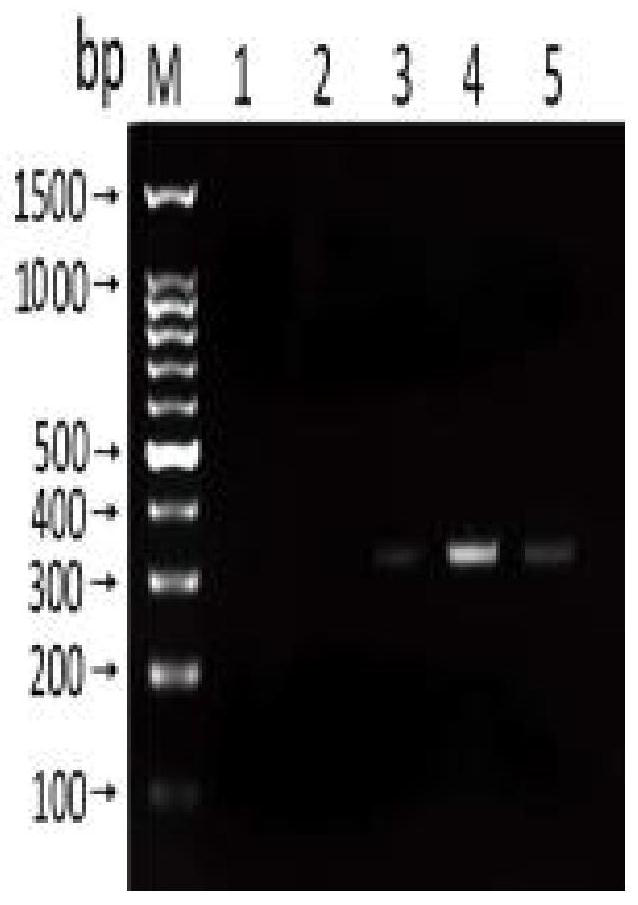
[0092] Sensitivity test: use Nanodroop 2000 to measure the concentration of Anisakis uteri DNA (100.5ng / μL), and dilute it with sterile water 10 times to 1ng / μL, 100pg / μL, 10pg / μL, 1pg / μL, 100fg / μL. RPA reactions were performed using diluted DNA at different concentrations as templates to determine the sensitivity of the method.
[0093] Specificity detection: respectively adopt the nematode uteri (100.5ng / μL), anisakis simplex (85.7ng / μL), anisakis peizii (60.3ng / μL), paraceca nematode (53.2ng / μL), Typical Anisakis (60.2 ng / μL) DNA was used as a template, and at the same time, Plasmocephalus ladenum (236.15 ng / μL), Clonorchis sinensis (200.6 ng / μL), Mesorchis orientalis (132.7 ng / μL), Echinostoma nematode (154.6ng / μL) was used as negative control and double distilled water was used as blank control, and RPA method was used for amplification. Add 1.5 μL of the genome of each worm body.
[0094] Sensitivity and sp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com