Nucleic acid sample treating method, sequencing method and kit
A sample and polynucleotide kinase technology, which is applied in the field of kits and nucleic acid sample processing, can solve the problems of not disclosing specific formulas, etc., and achieve the effects of saving operating time, simplifying operations, and facilitating industrialization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] The method for preparing the kit is exemplified below.
[0051] The kit contains tube 1, which is a mixed system with end repair function (also called End Prep Mix). In one case, tube 1 is a solution containing polynucleotide kinases and various DNA polymerases. In another case, tube 1 is a solution containing DNA fragmentation enzymes, polynucleotide kinases and various DNA polymerases.
[0052] The DNA polymerase can fill in the ends of the fragmented nucleic acid fragments. Here, preferably, the selected DNA polymerase has an amplification function, as well as repair and proofreading functions, such as selected from Taq DNA polymerase (referred to as Taq DNA polymerase for short). Enzyme) and one or both of Klenow fragment; polynucleotide kinase can be selected as T4 Polynucleotide Kinase (T4 PolynucleotideKinase).
[0053] In one case, the so-called DNA polymerase is a mixture of Taq enzyme and Klenow fragment. The concentration of Taq enzyme in tube 1 was (1-2) ...
Embodiment 2
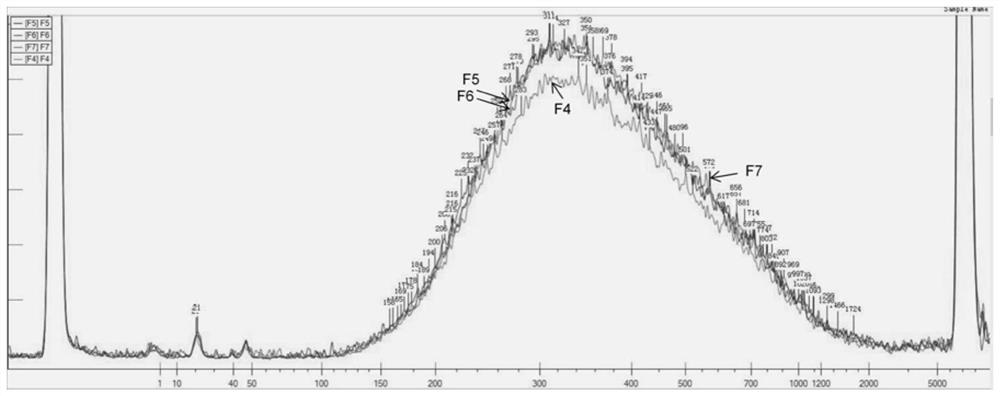
[0062] This example provides the application of the kit in Example 1 in library construction. A total of 4 parallel experiments were performed in this example, and the samples used were F4, F5, F6 and F7 genomes. The quality of the genomes needs to be preliminarily evaluated before the genomic DNA is fragmented to ensure that the genomes exist in the absence of metal ion chelators or other salts. solvent, and the genome integrity was checked by gel electrophoresis. The specific steps of library construction are as follows:
[0063] 1. Genomic DNA fragmentation, end repair and dA tailing
[0064] This step fragmented the DNA with blunt end-filling, phosphorylation at the 5' end and dA tailing at the 3' end.
[0065] Configure the reaction system in the PCR tube as shown in Table 1:
[0066] Table 1
[0067] H2O 14ul End Prep Mix 4ul DNA (25ng / ul) 2ul Total 20ul
[0068] Place the PCR tube in the PCR machine and set the reaction conditions a...
Embodiment 3
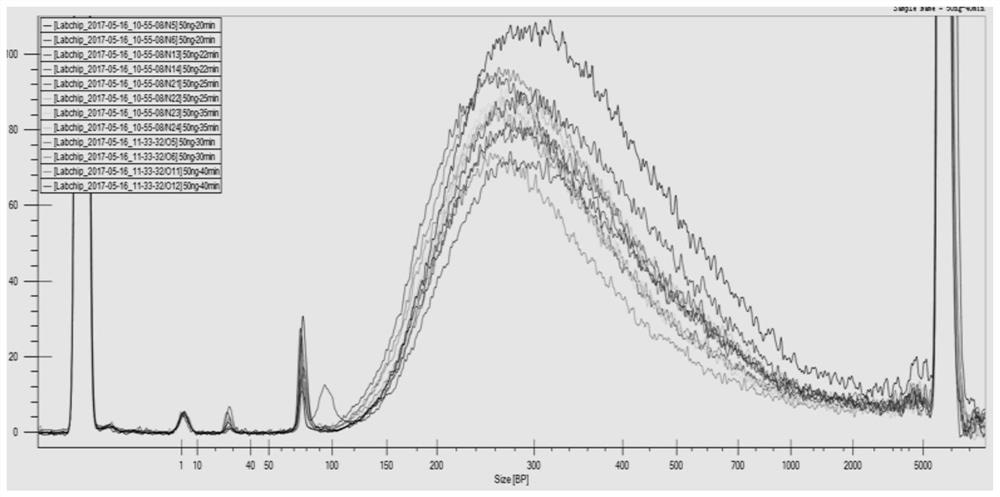
[0120] This example provides application 2 of the kit in Example 1 in library construction. A total of 12 parallel experiments were performed in this example, and the samples used were N5, N6, N13, N14, N21, N22, N23, N24, O5, O6, O11 and O12 genomes. Do a preliminary assessment to ensure that the genome is present in a solvent free of metal ion chelators or other salts, and to check for genome integrity by gel electrophoresis. The specific steps of library construction are as follows:
[0121] 1. DNA fragmentation, end repair and dA tailing
[0122] This step fragmented the DNA with blunt end-filling, phosphorylation at the 5' end and dA tailing at the 3' end.
[0123] Different samples have the same reaction system, as shown in Table 6, the reaction time at 32°C is different, 20min, 22min, 25min, 30min, 35min, 40min respectively.
[0124] The sample types and corresponding response times are as follows:
[0125] N5: 50ng, 32℃ for 20min; N6: 50ng, 32℃ for 20min;
[0126]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com