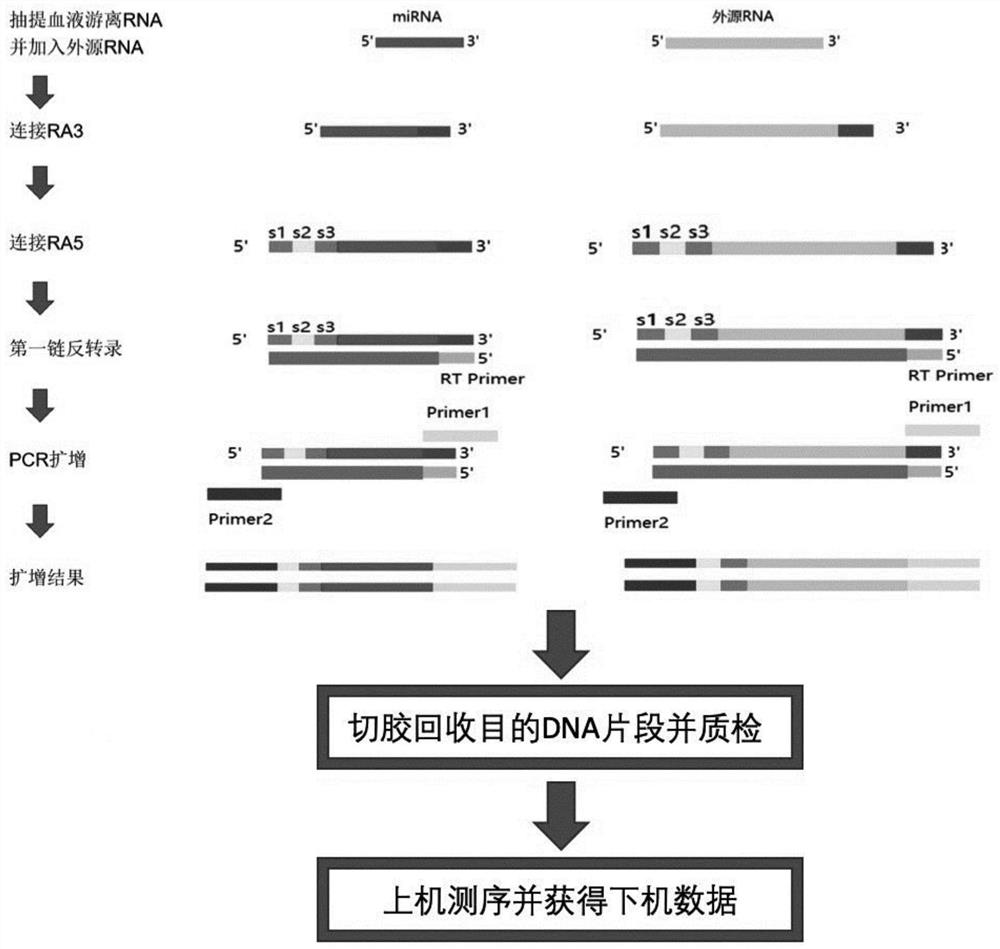
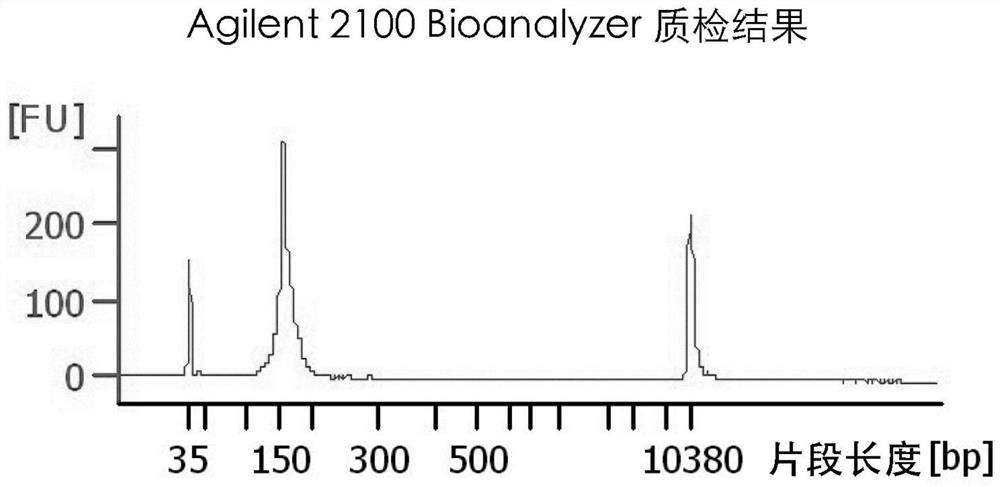
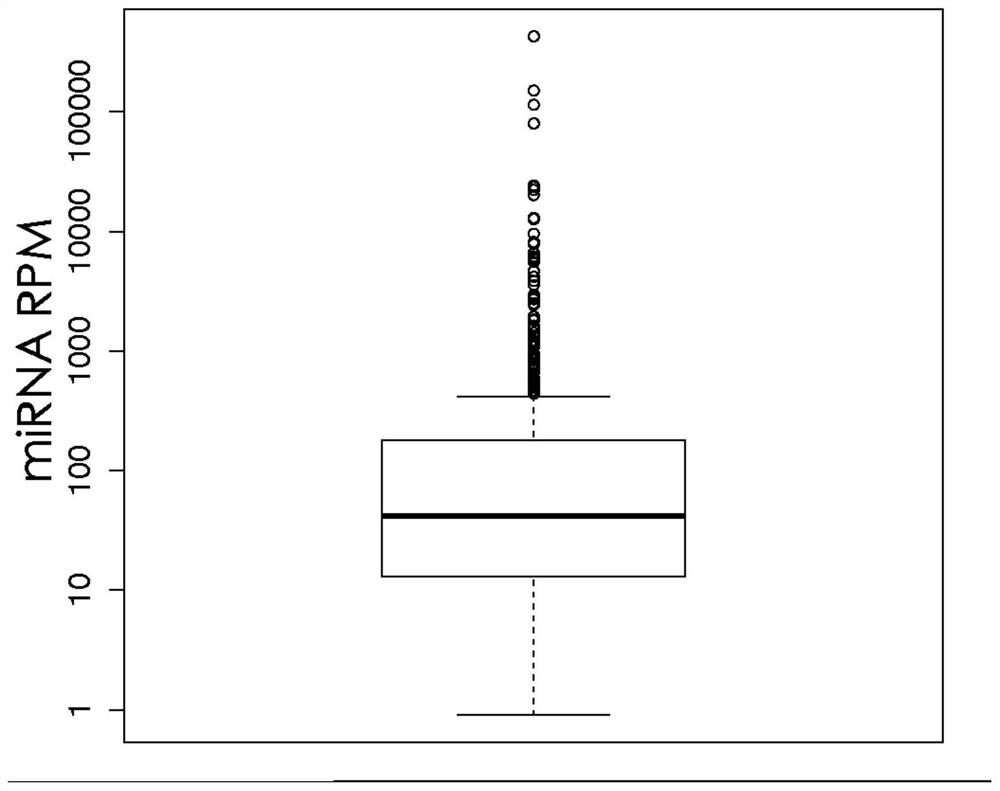
Method and kit for preparing library of free miRNAs in blood and quantitatively expressing free miRNAs
A kit and blood technology, which is applied in the field of free miRNA library preparation and expression quantification in blood, can solve the problems of the burden of the subjects, the library construction of the kit, and the failure to meet the requirements of the initial quantity of the library construction kit.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1: Obtaining a free miRNA sequencing library
[0063] For blood samples, the following method was used to obtain the free miRNA sequencing library:
[0064] (1) Collect 100 μl of human peripheral blood with a dry blood collection tube (non-anticoagulated), let it stand at 4°C for more than half an hour, then centrifuge at 400g for 10 minutes at 4°C to take the supernatant, and then centrifuge at 1800g for 10 minutes at 4°C to take the supernatant , obtain a serum sample (volume is about 30 μ l);
[0065] (2) Using miRNeasy Serum / Plasma Kit (Qiagen, product number 217184), extract a total of about 200pg of free RNA from serum samples, dilute it with ultrapure water (no DNase and RNase, the same below) to a total volume of 4 μl, and placed in a 200 μl thin-walled PCR tube.
[0066] (3) Add 1 μl of exogenous RNA to free RNA and mix well (that is, exogenous RNA and free RNA are mixed according to 50:1), the concentration of exogenous RNA is 10ng / μl, the length of ...
Embodiment 2
[0078] Example 2: Quantification of expression of miRNA
[0079] The obtained miRNA sequencing library was subjected to the following next-generation sequencing and data analysis to obtain the expression level RPM of miRNA:
[0080] (1) Send the miRNA sequencing library to the Illumina NextSeq 500 sequencing platform for sequencing, the sequencing read length is 75bp, and the sequencing mode is single-end sequencing, and obtain off-machine data;
[0081] (2) For the off-machine data, use FastQC, Cutadpat and Trimmomatic for data quality control and preprocessing (using default parameters) to obtain valid data with low-quality sequences and sequencing adapters removed; then the random tag sequence S2 in RA5 And the fixed base S3 is removed from the 5' end of the sequence of the valid data; then use the sequence comparison software Bowtie to re-align the obtained sequence to the human reference genome sequence (up to 1 base mismatch is allowed), and obtain the location at Locat...
Embodiment 3
[0084] Embodiment 3: the comparison of the method of the present application and prior art
[0085] In order to verify that the blood free miRNA library preparation and expression quantification method of the present application can produce reliable results, the miRNA sequencing results obtained by using the existing mainstream Illumina library construction kits were compared here.
[0086] From the same blood-donating individual in Example 1, at the same time and place, collect 30ml of peripheral blood with a dry blood collection tube (non-anticoagulated), and use the same method as in Example 1 to obtain a total amount of serum free RNA of about 1ug ; Subsequently, the control group: use the Illumina TruSeq Small RNA Library Preparation Kit (Catalog No.: RS-200-0012) for library preparation, see the kit operation instructions for the library construction method (the kit Contains no exogenous RNA, nor adapter sequences random tag sequence S2) ;After the sequencing library i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com