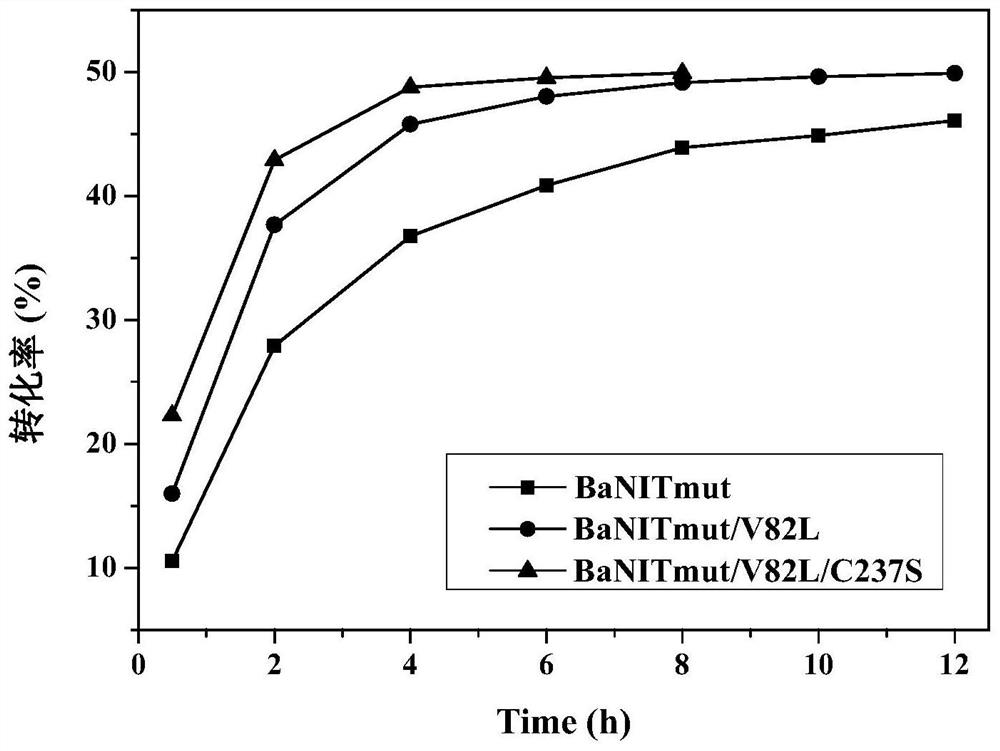
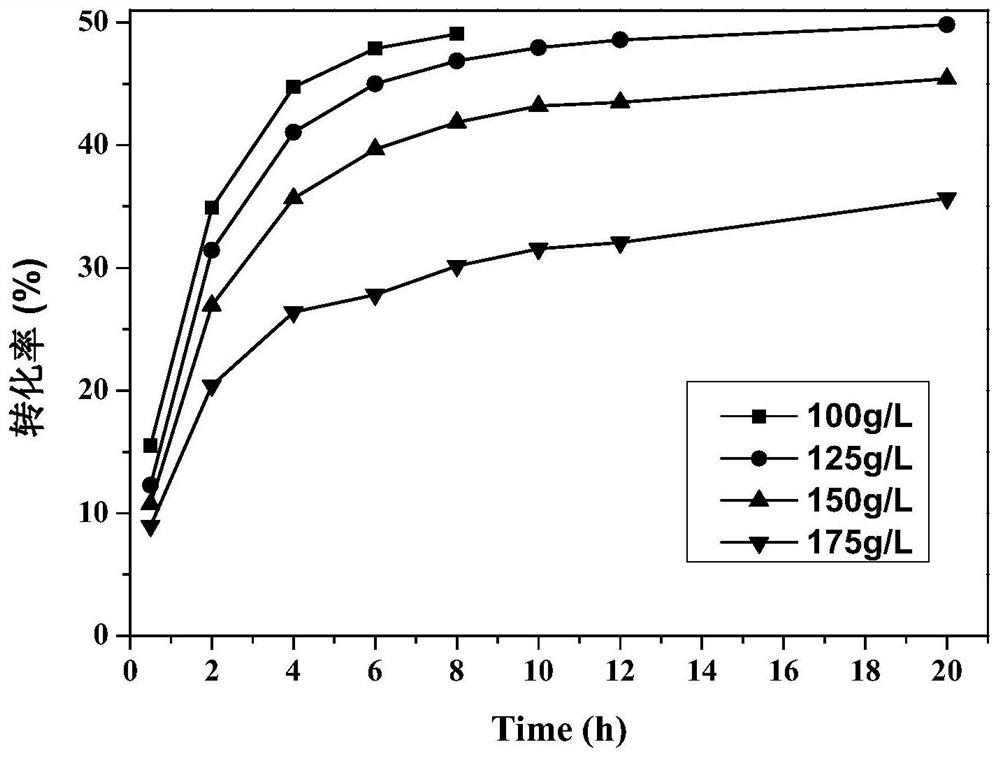
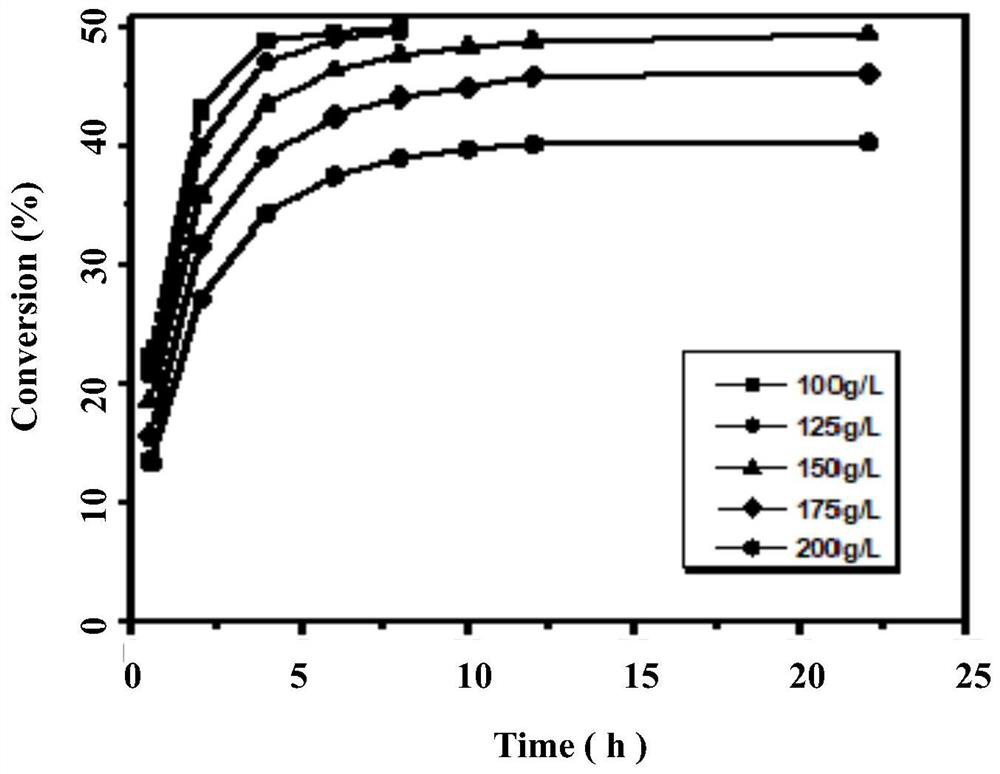
Nitrilase mutant with improved catalytic activity and reaction specificity and application
A technology of nitrilase and mutants, applied in the field of bioengineering, can solve the problems of production process and production cost increase, achieve the effects of reducing industrial production costs, improving nitrilase activity and reaction specificity, and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: Contain the construction of the recombinant escherichia coli of nitrilase mutant BaNITmut / V82L
[0045] In order to perform site-directed mutation at the 82nd position Val in the parental amino acid sequence, corresponding primers were designed, and the primer sequences are shown in Table 1. Using the recombinant plasmid pET28b-BaNITmut containing the target gene fragment as a template, its amino acid sequence is shown in SEQ ID NO.2, and its nucleotide sequence is shown in SEQ ID NO.1. According to the method of overlap extension PCR, the whole plasmid was carried out on the template Amplify.
[0046] Table 1: Primer Design Table
[0047] Primer name Primer sequence (5'to 3') V82L-Forward TATCGTTTCGGCATC GGTCTGGGTGTGCACAAC V82L-Reverse GTTGTGCACACCCCAGACCGATGCCGAAACGATA C237S-Forward TTCGTTCTGTCTGCTTCCCAGTTCTGCCGTCGT C237S-Reverse ACGACGGCAGAACTGGGAAGCAGACAGAACGAA
[0048] The PCR amplification system is (50 μ...
Embodiment 2
[0052] Example 2: Construction of recombinant E. coli containing nitrilase mutant BaNITmut / V82L / C237S
[0053] In order to construct the nitrilase double mutant BaNITmut / V82L / C237S, corresponding primers were designed, and the primer sequences are shown in Table 1.
[0054] Using the recombinant plasmid pET28b-BaNITmut / V82L constructed in Example 1 as a template, the construction method refers to Example 1 to obtain recombinant engineering bacteria E.coli BL21(DE3) / pET28b-BaNITmut / V82L / C237S, the amino acid sequence of which is as SEQ ID NO. 6, and the nucleotide sequence is shown in SEQ ID NO.5.
Embodiment 3
[0055] Example 3: Induced expression of recombinant Escherichia coli containing nitrilase mutants
[0056] Inoculate the recombinant Escherichia coli of the parental nitrilase BaNITmut and the mutants BaNITmut / V82L and BaNITmut / V82L / C237S obtained in Examples 1 and 2 into LB liquid medium containing 50 μg / mL kanamycin, and cultivate overnight at 37°C , then inoculated into LB medium containing 50 μg / mL kanamycin with 2% inoculum size (v / v), cultivated to thalline concentration OD at 37° C., 150 rpm 600 = about 0.6, add IPTG with a final concentration of 0.1mM, induce culture at 28°C for 12h, then centrifuge at 4°C and 12000rpm for 10min to collect the bacterial cells, wash the wet bacterial cells with 0.85% normal saline, and store them at -20°C for later use (i.e. static cells for hydrolysis reactions).
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com