Biological analysis method based on quantum dot selective recognition reaction and application thereof
A biological analysis and selective technology, applied in the field of biomedical diagnostic analysis, can solve the problems of limited analytical sensitivity of analytical methods, and achieve the effect of improving analytical sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] This example provides the synthesis method of CdTe QDs.
[0030] The CdTe QDs of this example were synthesized by a one-pot method, as follows:
[0031] First, 0.5mmol CdCl 2 and 0.20 g of trisodium citrate were dissolved in 50 ml of water, and 52 μL of mercaptopropionic acid (MPA) was added to the above solution. Using NaOH solution, the pH of the above mixture solution was adjusted to 10.5. Then, 0.1mmolNa 2 TeO 3 and 50mg KBH 4 Add it to the above solution, and reflux for 1 hour until the solution turns red, showing strong red fluorescence under the irradiation of ultraviolet light. Finally, the CdTe QDs solution was purified by precipitation (using n-propanol) and centrifugation (11000 rpm, 30 min). The MPA-CdTe QDs synthesized above were stored at 4 °C before use.
Embodiment 2
[0033] This example provides the preparation methods and selective cation exchange reaction conditions for different DNA template CuNPs. details as follows:
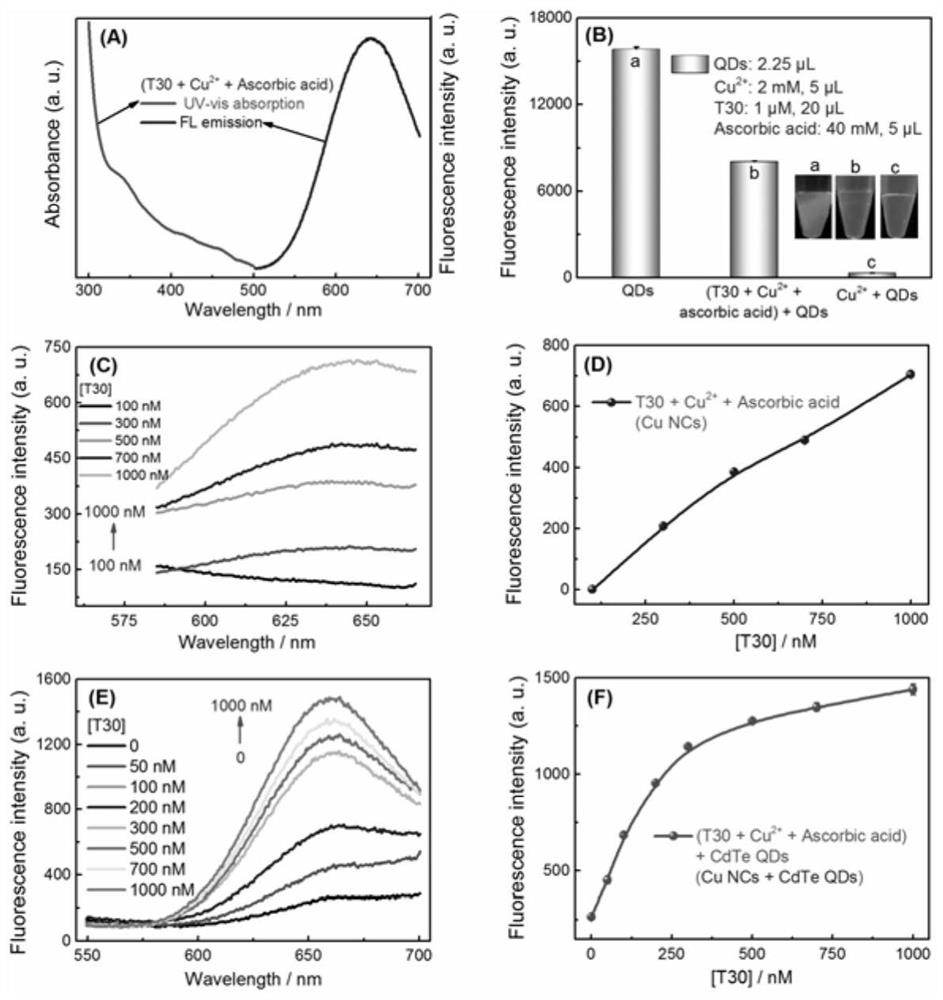
[0034] Shake 70 microliters of 10mmol / L 3-(N-morpholine) propanesulfonic acid (MOPS) buffer solution, 20 microliters of DNA strands (poly T chain, dsDNA) with different concentrations and mix well, add 40mmol / L 5 microliters of ascorbic acid ; shake for 30 seconds; then add 5 microliters of 2mmol / L Cu 2+ , oscillating for 30 seconds, and standing for 3 minutes at room temperature to react; finally, use a molecular fluorescence spectrophotometer to detect the fluorescence signal.
[0035] Add CdTe QDs to the Cu NPs solution of different DNA strand templates above, and let it stand for 3 minutes at room temperature. Subsequently, the fluorescence signal changes were detected using a molecular fluorescence spectrophotometer.
[0036] In order to elaborate on the effect of QDs on Cu 2+ and the selective recognition pheno...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com