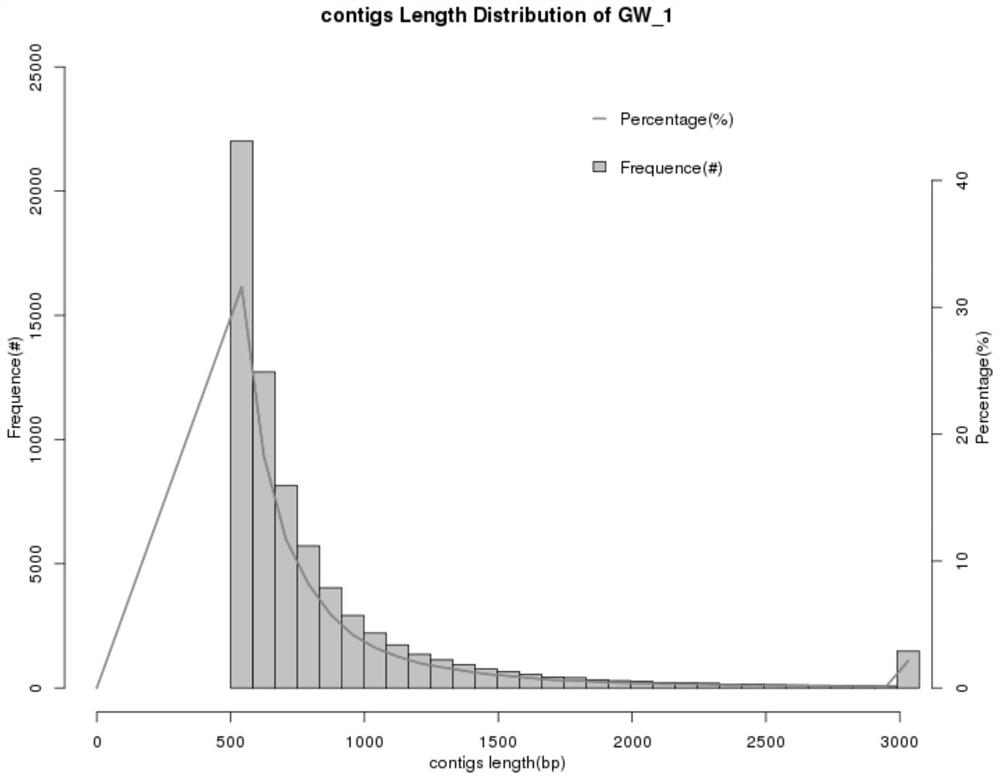
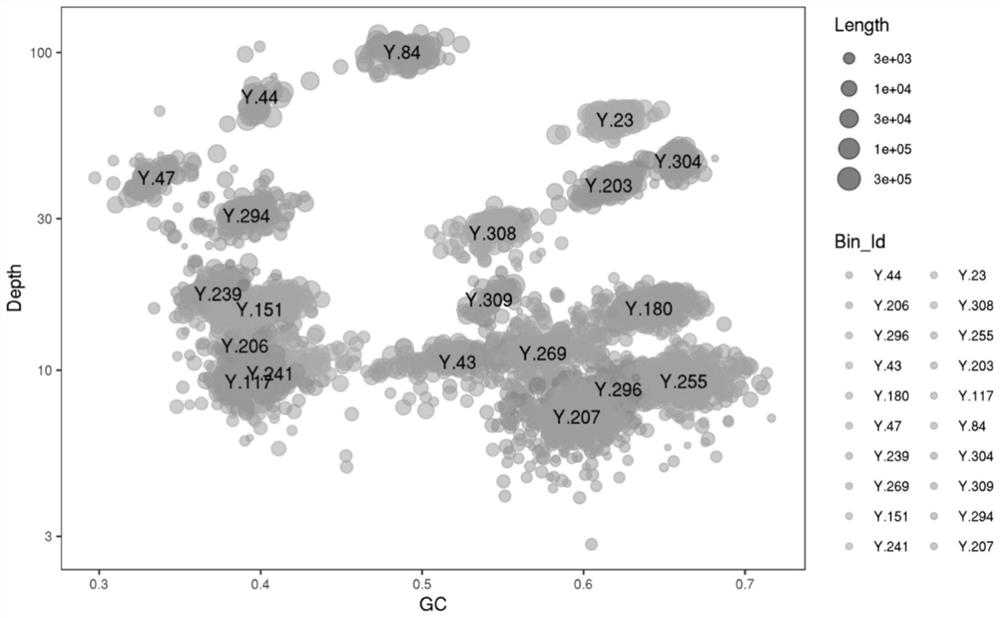
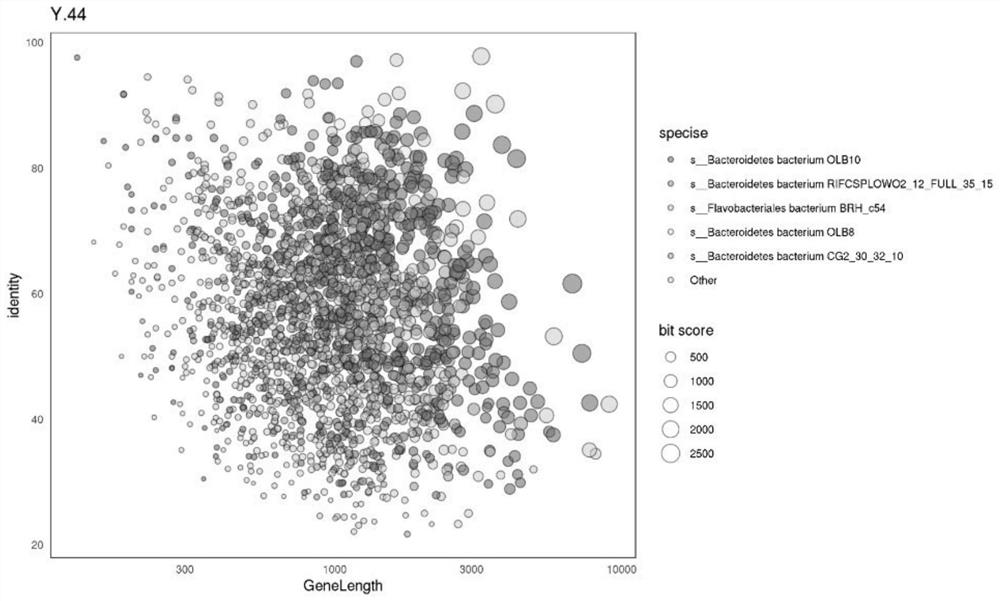
Optimized method for analyzing microbial communities through metagenome binning
A metagenomic and microbiota technology, applied in the field of high-throughput sequencing technology and bioinformatics analysis of sequencing data, can solve problems such as difficulty in providing bin gene annotations, missing target new species, and lack of intuitive understanding by users
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0172] Example 1 (Assembly Strategy 2)
[0173] Non-natural environmental samples, four cases of different human intestinal samples (a total of about 6Gb data) in the assembled condition, the prior art current sample intestinal preferably assembled effect Document Han M, Yang P, Zhong C, et al .The Human GutVirome in Hypertension [J] .Frontiers in Microbiology, 2018,9: the assembly of theHuman Gut Metagenomic Data analyzing section Document N50 is 4152bp, and reference Examples of the present embodiment is assembled using the parameters 27,37,47 kmer , 57,67,77,87,97,107,117,127, assembled embodiment of the present embodiment N50 higher than the prior art, better assembly.
[0174] quantity Total length Average length N50 N90 The maximum length Minimum length GC Sample 1 27273 87611145 3212.38 4785 1263 371303 1000 48.54% Sample 2 26592 88292244 3320.26 4967 1329 124147 1000 57.51% Sample 3 17137 58929791 3438.75 5418 1315 277887...
Embodiment 2
[0176] Second Embodiment (policy assembly 4):
[0177] The prior art literature (Zhang M, Pan L, Huang F, et al.Metagenomic analysis ofcomposition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments underdifferent culture modes [J] .Aquaculture, 2019 , 506: 280-293) in three cases the natural environment of water samples i.e. samples (GW, HE, HW) data, the process of the present invention to art methods, existing parameters, and literature data comparison, the following results:
[0178] (1) GW water, described in the literature a total length of 288 496, N50 is 954, the sample data for comparison to the prior art parameters and techniques of the present invention, the result data are as follows, techniques N50 present invention is 1027, the assembly was better than art parameters.
[0179]
[0180]
[0181] (2) HE water, described in the literature a total length of 356 232, N50 is 914, the sample...
Embodiment 3
[0185] Third Embodiment (3 assembled policy)
[0186] Natural environmental samples - sediment samples three cases, the amount of data. 6G single embodiment, mixing assembly reads the set of samples, better mixing assembly.
[0187] quantity Total length Average length N50 N90 The maximum length Minimum length GC Sample 1 177935 289272894 1625.72 2560 618 755937 500 62.61% Sample 2 177055 287062202 1621.32 2559 617 755937 500 62.63% Sample 3 174799 283115279 1619.66 2579 616 755937 500 62.64% Mix assembly 325995 578768707 1775.39 3070 646 1275998 500 62.52%
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com