PCR primer and quantitative detection method for rapidly detecting strawberry colletotrichum gloeosporioides
A technology for the detection of glyospora anthracnose bacteria, applied in biochemical equipment and methods, microbe measurement/inspection, DNA/RNA fragments, etc., can solve problems such as high information error rate, difficulty in detection, and decline in real quality, and achieve High amplification efficiency, good specificity, time-saving and cost-saving effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1 A kind of PCR rapid detection method of G. fragariae anthracnose
[0033] 1. Primer design
[0034]In order to detect the compound species of G. glyospora anthracnose on strawberries in the field in my country as far as possible, the cutinase gene sequences of 35 strains of G. anthracis gloeosporum collected from different regions of the country were respectively amplified and sequenced. After sequence comparison, the selected It has a common nucleic acid sequence at the 3' end of the gene, as shown in SEQ ID NO.1.
[0035] The selected conserved sequence is located at the 3' end of the cutinase gene, which is unique to the physiological race of G. fragariae. The cutinase encoded by this gene is involved in assisting the invasion process of anthracnose. It is speculated that it may be related to the high prevalence of G. It is related to pathogenicity, and the expansion and identification of specialized physiological races in different regions of my country ...
Embodiment 2
[0056] Example 2 A method for absolute quantification of the biomass of strawberry anthracnose pathogenic bacteria Gleospora anthracnose
[0057] 1. Establish a standard curve
[0058] The standard plasmid pGEM-T-cut series after 10-fold isocratic dilution was used as a template, and the concentrations were 3.28×10 1 copies / μL, 3.28×10 2 copies / μL, 3.28×10 3 copies / μL, 3.28×10 4 copies / μL, 3.28×10 5 copies / μL, use primers Cfcut1 and Cfcut2 to carry out real-time fluorescent quantitative PCR amplification, obtain the cycle number Cq value of the corresponding standard, use the obtained cycle number Cq value and the corresponding plasmid template copy number to plot, and obtain the standard curve equation Y= -3.3348 (log 10 X)+34.726, Y is the cycle number Cq value, X is the initial copy number, correlation coefficient R 2 The value is 0.9992, and the primer amplification efficiency is 99.47%.
[0059] Among them, the real-time fluorescent quantitative PCR reaction syste...
Embodiment 3
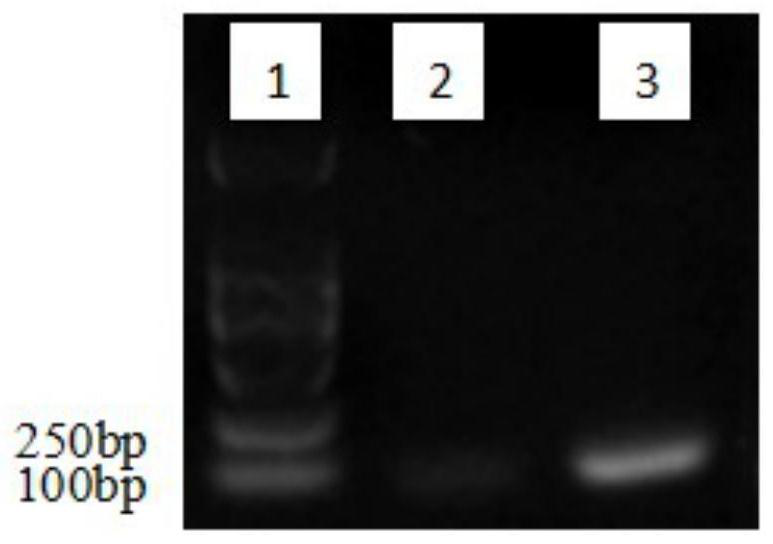
[0065] Four physiological races of G. spp. C.aenigma, C. fructicola, C. siamense and C. gloeosporioide, C. nymphaeae and other three common pathogenic fungi in the field, Botrytiscinerea, Sphaerotheca macularis and Pestalotiopsis clavispora Genomic DNA of healthy strawberries and healthy strawberries were used as samples for detection.
[0066] 1. Extraction of fungal genomic DNA:
[0067] (1) Take 100 mg of fresh mycelia into a centrifuge tube, quickly add 600 μL of FG1 buffer solution, shake vigorously to fully disperse the mycelia, incubate at 65°C for 10 minutes, and mix by inverting twice during the period;
[0068] (2) Add 140 μL of FG2 buffer, shake and mix well, centrifuge at ≥10,000 g for 10 minutes, carefully absorb about 600 μL of the supernatant and transfer it to a new centrifuge tube, add 0.7 times the volume of isopropanol and mix well;
[0069] (3) Discard the supernatant after centrifugation at 10,000 g for 2 minutes, add 300 μL sterile deionized water prehea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com