Cold-adapted ribonuclease R and coding gene and application thereof
A technology of ribonuclease and coding genes, which can be used in applications, genetic engineering, plant genetic improvement, etc., and can solve problems such as poor stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Cloning and sequencing analysis of ribonuclease R gene.
[0019] Antarctic microbes Psychrobacter sp. ANT206 was activated in 2216E liquid medium, cultured to the middle and late logarithmic growth phase (about 4d), and the total gene DNA of the strain was extracted by combining CTAB method and phenol-chloroform extraction method. Using the extracted total DNA as a template, PCR was performed using degenerate primers.
[0020] Upstream primer: 5'- ATGANGGAATGATCRACGCCT -3'
[0021] Downstream primer: 5'- ANGTTTGGATCATTNACTCAT -3'
[0022] Amplification conditions were: denaturation at 94°C for 1 min, annealing at 54°C for 1 min, extension at 72°C for 90 s, and 30 cycles. Then, agarose gel electrophoresis was used to detect the band containing the target gene cold-adaptive ribonuclease R and sequenced. By analyzing the sequencing results, a gene with a complete reading frame sequence of 2 313 bp in full length was obtained. The nucleotide sequence is shown in SEQ ID ...
Embodiment 2
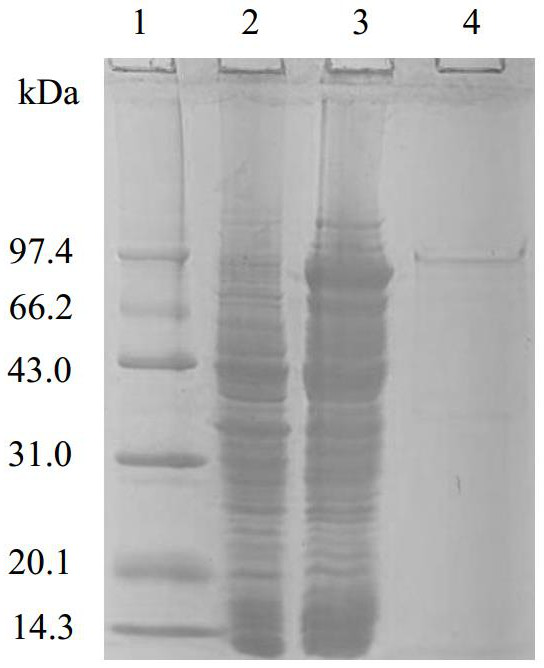
[0024] Expression and purification of ribonuclease R gene
[0025] According to the determined full-length sequence of cold-adapted ribonuclease R, primers containing restriction sites were redesigned.
[0026] Upstream primer: 5’-ACT GGATCC ATGTCAAACCAAGATC -3'
[0027] Downstream primer: 5’-TAC CTCGAG CGCTCTTTTTACTACT -3'
[0028] The lines are Bam Hi, xho I restriction site.
[0029] The cold-adapted ribonuclease R gene and pET-28a (+) double-digested gel recovery product were connected in proportion with T4 ligase to construct a recombinant expression vector. Transform recombinant expression vectors into competent cells E. coli In BL21, positive clone screening and enzyme digestion verification were carried out.
[0030] The recombinant strain obtained by screening was induced to express by IPTG. Inoculate the recombinant bacteria into LB medium and cultivate to OD at 32-40°C 600 0.6-0.8, IPTG was added to the medium to a final concentration of 0.5-1.5 mM, ...
Embodiment 3
[0032] Study on the Enzymatic Properties of Recombinant Cold Adaptive Ribonuclease R
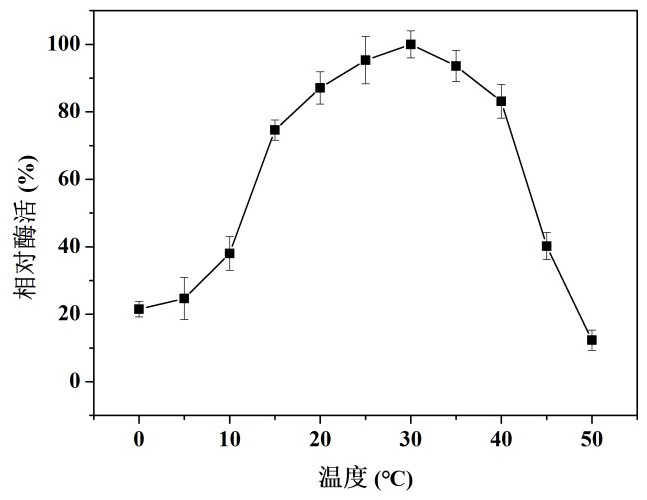
[0033] (1) Determination of the optimum reaction temperature: Under the conditions of different temperatures (0 ~ 50 ℃) and pH 6.0, the enzymatic activity of cold-adapted ribonuclease R was determined, and the highest enzymatic activity was set as 100%, and the activity at other temperatures was The ratio of the enzyme activity to the highest enzyme activity is the relative enzyme activity of the enzyme solution to be tested at this temperature. The result is as figure 2 As shown, the results show that the optimum action temperature of the enzyme is between 28 ~ 32 ℃, and the enzyme activity is maintained at more than 85% of the highest enzyme activity at 20 ~ 35 ℃. When the temperature exceeds 35 ℃, with the temperature As the temperature increases, the enzyme activity gradually decreases, and when the temperature reaches above 50 °C, the enzyme activity is basically lost.
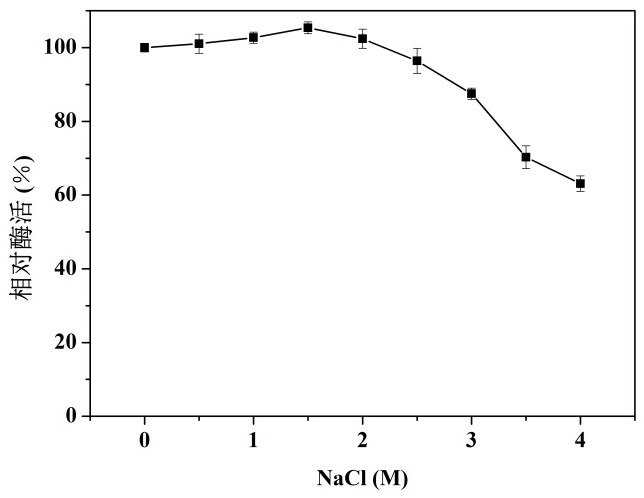
[0034] (2) T...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com