An Annotation Method for Variant Sequences
A variation and sequence technology, applied in the field of annotation of variation sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0070] (1) Determine the variant sequence information (call variants)
[0071] (1.1) Obtaining variant sequences
[0072] Using probe capture technology, next-generation sequencing of human whole exons is used to obtain the sequence to be analyzed. Use variant sequence analysis software (GATK, https: / / gatk.broadinstitute.org / hc / en-us) to compare the sequence to be analyzed with the reference genome to obtain call variants.
[0073] (1.2) Integrate reference sequence information
[0074] Obtain the hg19 reference genome sequence and the hg19 reference genome annotation file, which includes gene name, transcript name, physical location, positive and negative strands, information about each element (elements include UTR, Intron, CDS), etc. Among them, the download address of the hg19 reference genome sequence is ftp: / / hgdownload.soe.ucsc.edu / goldenPath / hg19 / bigZips / hg19.fa.gz; the access address of the hg19 reference genome annotation file is: ftp: / / hgdownload. soe.ucsc.edu / go...
example 1
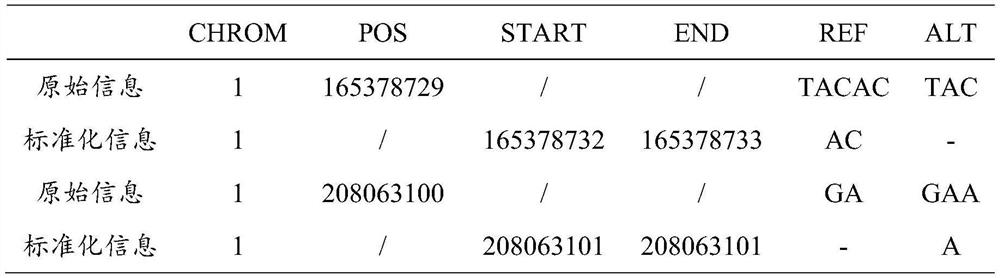
[0130] Mutation site: A to G at position 69511 of chromosome 1 (step 1.3 standardized mutation information, 1:69511:69511:A:G)
[0131] Amino acid sequence variant annotation results:
[0132] Comparative example: OR4F5:NM_001005484:exon1:c.421A>G:p.T141A
[0133] Example: OR4F5:NM_001005484:exon1:c.421A>G:p.Thr141Ala
[0134] The annotation results are consistent, but the one-letter abbreviation of amino acid used by ANNOVAR in the comparative example does not conform to the specification.
[0135] Nucleic acid sequence variant annotation results:
[0136] Comparative ratio: Symbol: OR4F5
[0137] Example: Symbol: OR4F5, EntrezID: 79501
[0138] The annotation results are consistent, but there is no EntrezID information in ANNOVAR in the control example.
[0139] Functional area notes: All are exonic; results are consistent.
[0140] Variation type annotation: All are nonsynonymous SNVs; the results are consistent.
example 2
[0142] Variation site: G deletion at position 70176769 of chromosome 9 (step 1.3 normalized variation information, 9:70176769:70176769:G:-)
[0143] Nucleic acid and amino acid sequence variation annotation results:
[0144] Comparative example: FOXD4L5:NM_001126334:exon1:c.1215delC:p.W406Gfs*21
[0145] Example: FOXD4L5:NM_001126334:exon1:c.1215_1215del:p.Trp406Glyfs
[0146] The annotation results are consistent, but the example shows the deletion of the start and stop sites, and the three-letter amino acid is used.
[0147] Functional area notes: All are exonic; results are consistent.
[0148] Mutation type annotation:
[0149] Comparative example: frameshift deletion
[0150] Example: del_frameshift_stoploss
[0151] In the example, the stoploss information is successfully annotated.
[0152] Genetic information:
[0153] Comparative ratio: Symbol: FOXD4L5
[0154] Example: Symbol: FOXD4L5, EntrezID: 653427
[0155] ANNOVAR has no EntrezID information in the comp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com