Capture probes for large genomic rearrangement detection based on capture sequencing, kit and detection method
A technology for capture probes and large fragments, applied in the field of capture probes, can solve problems such as limited amplicons, false positives, and reduced amplification efficiency, and achieve the effects of improving detection performance, improving detection capabilities, and reducing purity requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] Example 1 Preparation and sequencing of tissue samples
[0092] Use the QIAamp DNA FFPE Tissue Kit (50) (product number: 56404) to extract the DNA of the tissue sample, and then use the ultrasonic breaker Covaris Focused-ultrasonicator (product number: S220) to break the extracted DNA into fragments of about 200bp, and then The end of the fragment is repaired and phosphorylated, and then deoxyadenine (A) is added to the 3' end of the fragment, and then the linker is connected. The treated fragments were amplified using ProFlex 3x 32well PCR system, and the amplified products were hybridized with probes (SEQ ID NO: 1-584). After hybridization, the fragments were specifically eluted, and then PCR amplified. The amplified products were then quantified and fragment length distribution determined. After qualifying, use Illumina's NovaSeq6000System for sequencing, and the sequencing depth is not lower than 500X.
Embodiment 2
[0093] Example 2 Preparation and sequencing of blood samples
[0094] Use the QIAamp DNA Mini Kit (250) (product number: 51306) to extract nuclear DNA from peripheral blood leukocytes, and use the ultrasonic breaker Covaris Focused-ultrasonicator (product number: S220) to break the extracted DNA into about 200bp as in tissue samples The fragment, then repair and phosphorylate the end of the fragment, and then add deoxyadenine (A) to the 3' end of the fragment, and then connect the adapter. The treated fragments were amplified using ProFlex 3x 32well PCR system, and the amplified products were hybridized with probes (SEQ ID NO: 1-584). After hybridization, the fragments were specifically eluted, and then PCR amplified. The amplified products were then quantified and fragment length distribution determined. After qualifying, use Illumina's NovaSeq 6000System for sequencing, and the sequencing depth is not lower than 500X.
Embodiment 3
[0095] Embodiment 3 bioinformatics analysis
[0096] 1 Sequencing data preprocessing and quality control
[0097] Use bcl2fastq v2.19.0 software to convert the sequencing off-machine file (bcl format) to sequence file (fastq format), and then use fastp v0.20.0 software to perform quality control (QC) and filter (filter) on the sequence file to remove low-quality sequences , and then use bwa v0.7.12 software to align high-quality clean sequences to the human reference genome (GRCh37), generate an alignment file (bam format), and use picard v1.8.0_221 to sort and deduplicate the file (PCRduplication ) processing, using the bcftools v0.1.19 software package to detect the genotypes and corresponding depth information (depth) of all sites in the target region, and using the local Python script to calculate the GC content of the probe region.
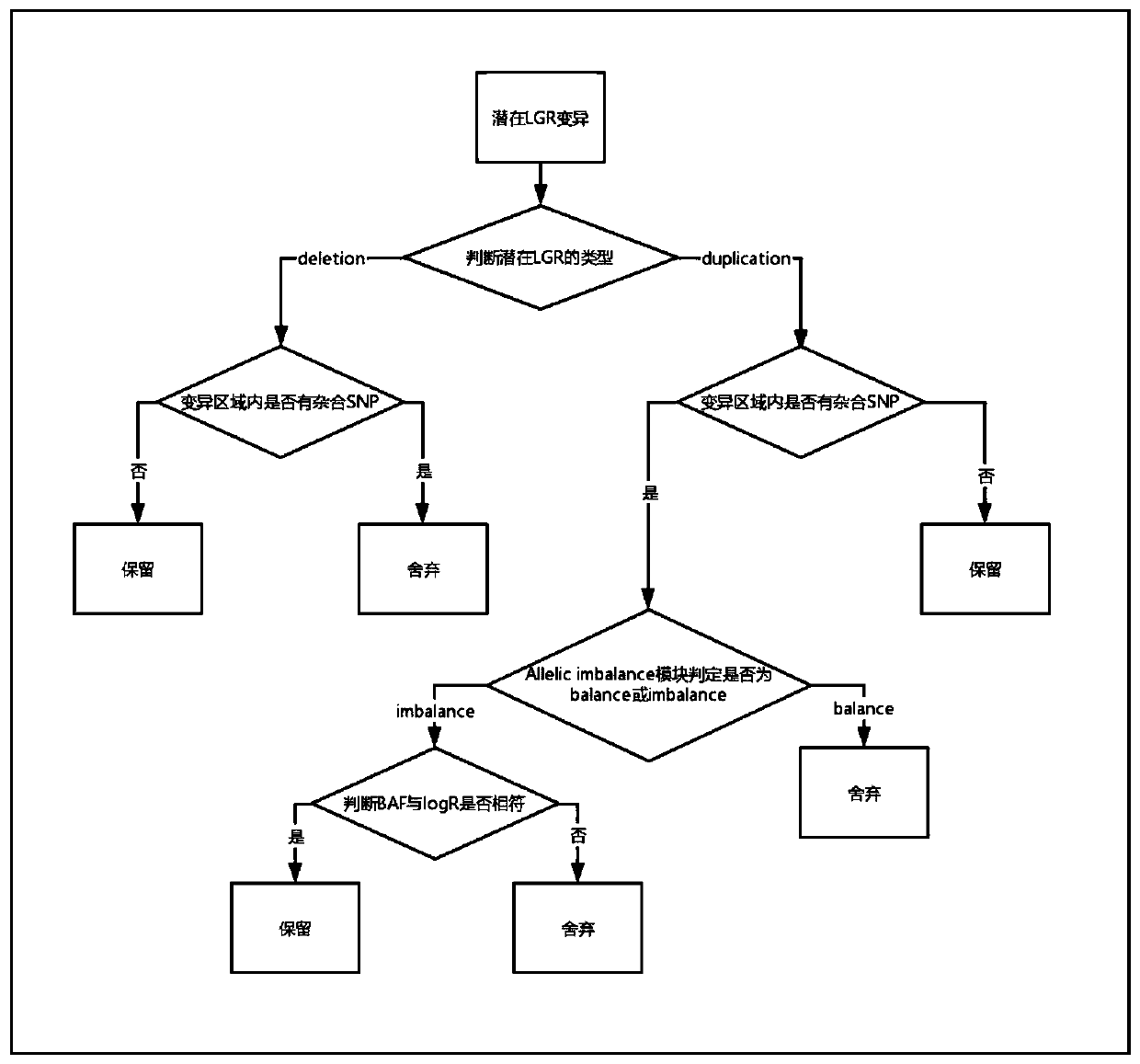
[0098] 2 Preliminary determination of potential LGR variants
[0099] (1) Reads depth determination: Count the number of sequences (reads ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com