Fluorescent quantitative PCR (polymerase chain reaction) method for detecting enterocytozoon hepatopenaei of prawns and corresponding kit
A fluorescence quantification and kit technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low detection sensitivity, long detection time, and high false positives, and achieve high sensitivity, convenient operation, and rigorous results. and accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 detection primer amplification standard curve
[0059] 1. Microbial culture
[0060] Use LB medium as the culture medium for the E. hepatica plasmid of prawns, and it grows well in the plate with pH 7.0~7.4. The diameter of the colony on the plate is 2mm, round, smooth and transparent. The vigorously growing microbial samples were selected for subsequent experiments.
[0061] 2. Genomic DNA Extraction
[0062] Genomic DNA was extracted according to the instructions of Qiagen's QIAamp DNA Mini Kit.
[0063] 3. Production of standard plasmids for amplified products
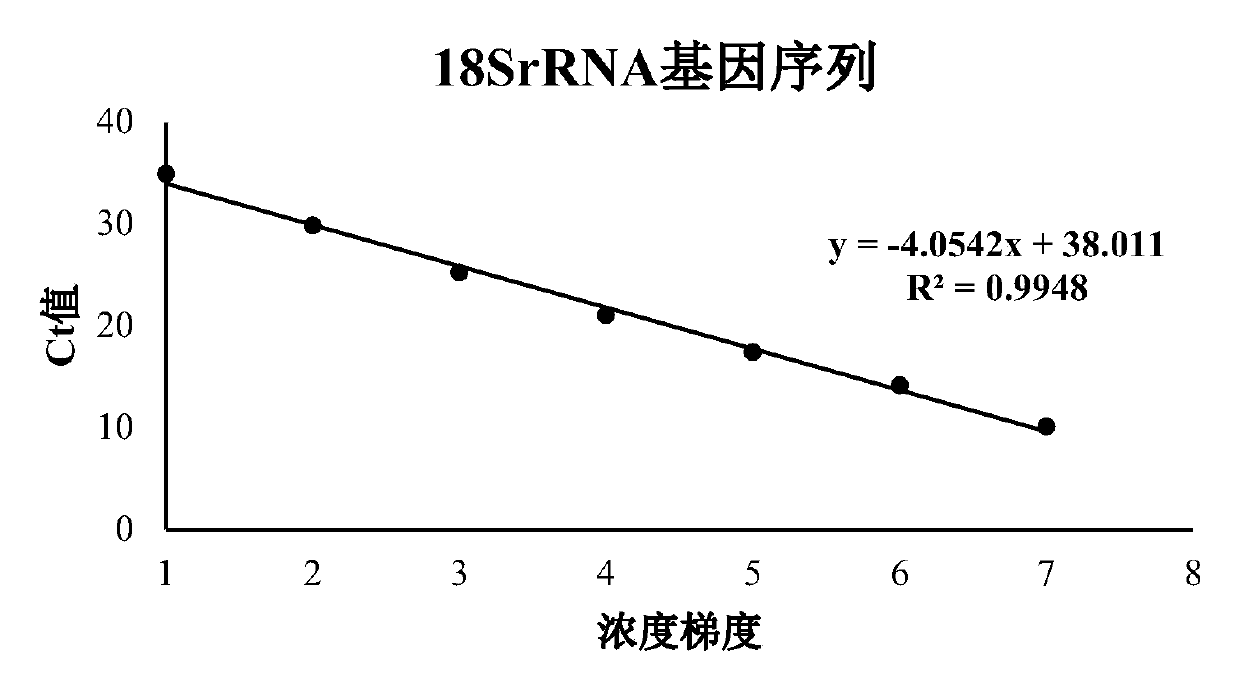
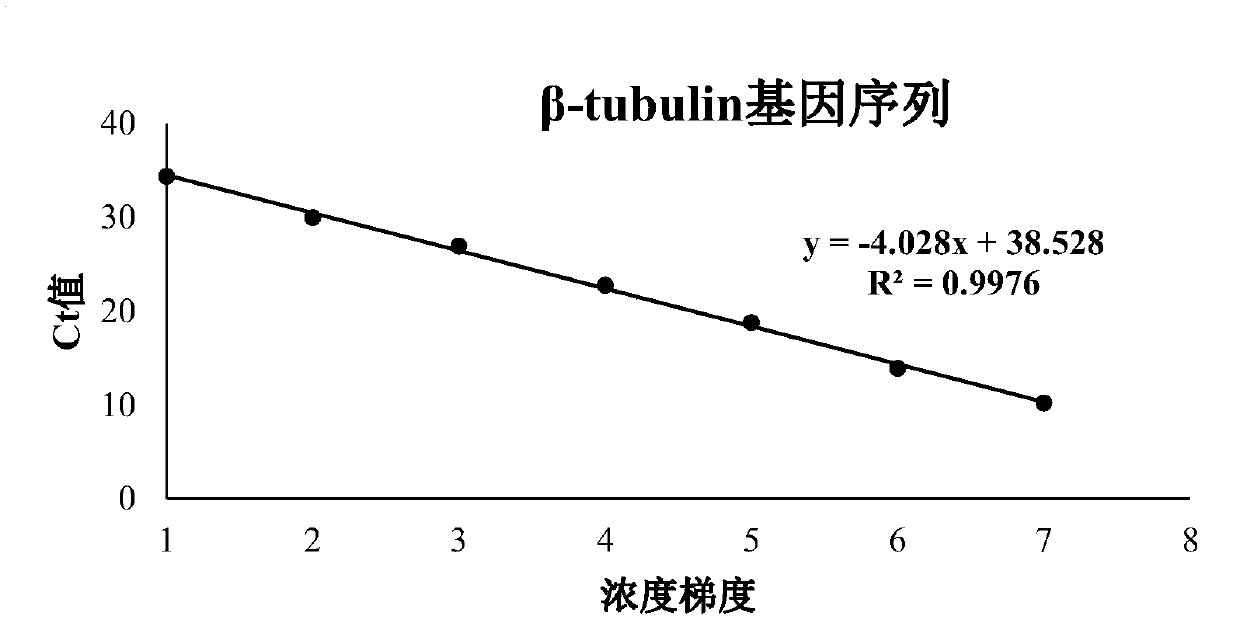
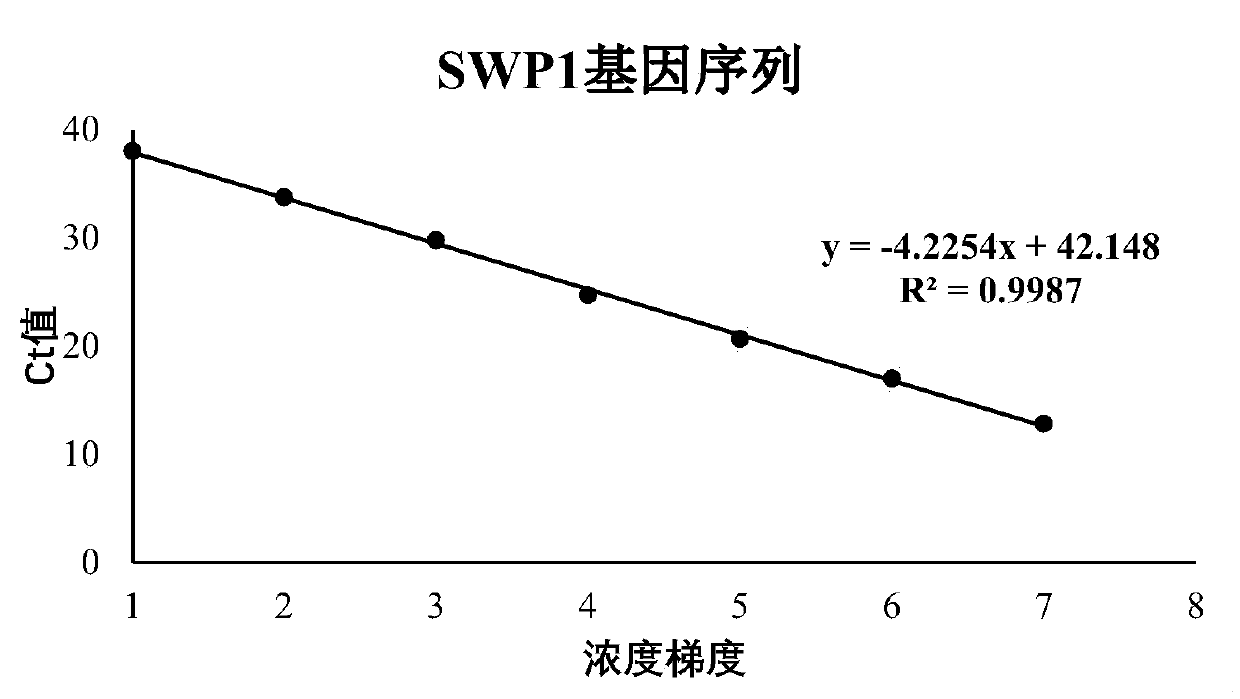
[0064] Using the DNA extracted from prawn Enterospora hepatica, amplified with the primers of 18SrRNA gene, β-tubulin gene and SWP1 gene respectively, after adding A treatment, the amplified product was ligated into the T vector using T4 ligase, and passed After transduction of competent cells, the plasmid was amplified on a large scale. The primer sequences of the three genes are shown in ...
Embodiment 2
[0083] Example 2 Plasmid Repeatability Experiment
[0084] 1. Microbial culture
[0085] With embodiment 1.
[0086] 2. Genomic DNA Extraction
[0087] With embodiment 1.
[0088] 2. qPCR detection of 3 target genes
[0089] Carry out qPCR amplification of the sample according to the instructions of TaKaRa Taq PCR Mix, and obtain the Ct value reading corresponding to each pair of primers. The qPCR reaction in this experiment was set at two concentrations of 1×10 6 fg / μL and 1×10 5 fg / μL, 4 biological replicates and 5 technical replicates were set up for each concentration, and the qPCR reaction system and amplification reaction conditions are shown in Table 7.
[0090] Table 7
[0091]
[0092] 4. Result Analysis
[0093] In this example, the correlation coefficient and reliability of the amplification curves of the three genes were analyzed, as well as the amplification efficiencies at two concentrations. The measured CT values were all less than 35 cycles, and a...
Embodiment 3
[0101] Embodiment 3 Positive and negative sample detection situation
[0102] 1. Microbial culture
[0103] With embodiment 1.
[0104] 2. Genomic DNA Extraction
[0105] With embodiment 1.
[0106] 3. qPCR detection of 3 target genes
[0107] Carry out qPCR amplification of the sample according to the instructions of TaKaRa Taq PCR Mix, and obtain the Ct value reading corresponding to each pair of primers. Each qPCR reaction was set up with 3 biological repetitions and 3 technical repetitions, and the qPCR reaction system and amplification reaction conditions were the same as in Example 2.
[0108] 4. Result Analysis
[0109] as shown in Table 11 and Figure 10As shown, the qPCR amplification results of 3 pairs of primers showed that E. hepatica had three positive results of 18SrRNA, β-tubulin gene and SWP1 gene, while other viruses or bacterial genera showed negative results. Therefore, for E. hepatica and other viruses or fungi, the detection of 18SrRNA, β-tubulin ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com