High-fault-tolerance genome complex structure variation detection method based on filtering strategy
A complex structure and mutation detection technology, applied in genomics, proteomics, instruments, etc., can solve problems such as interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
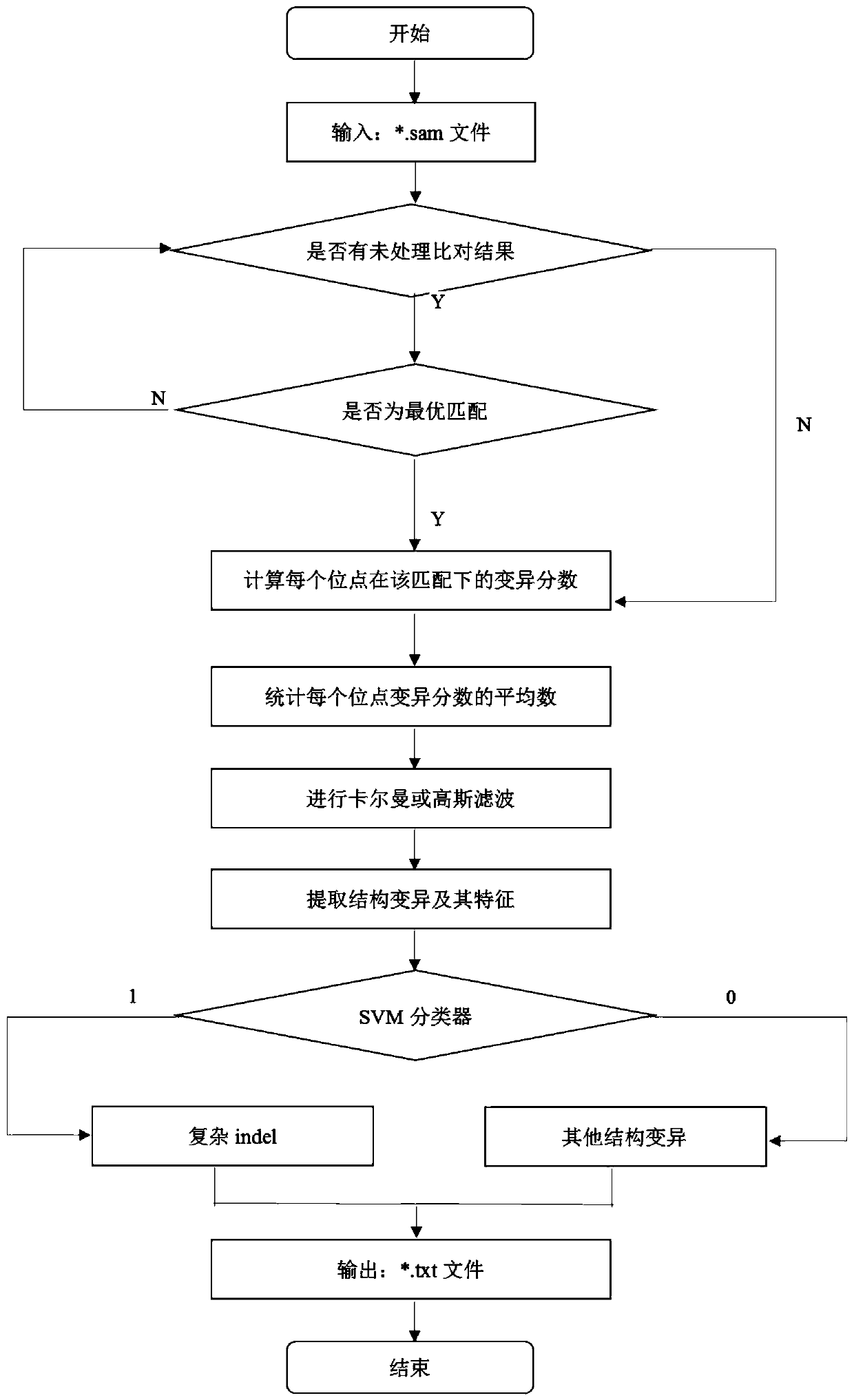
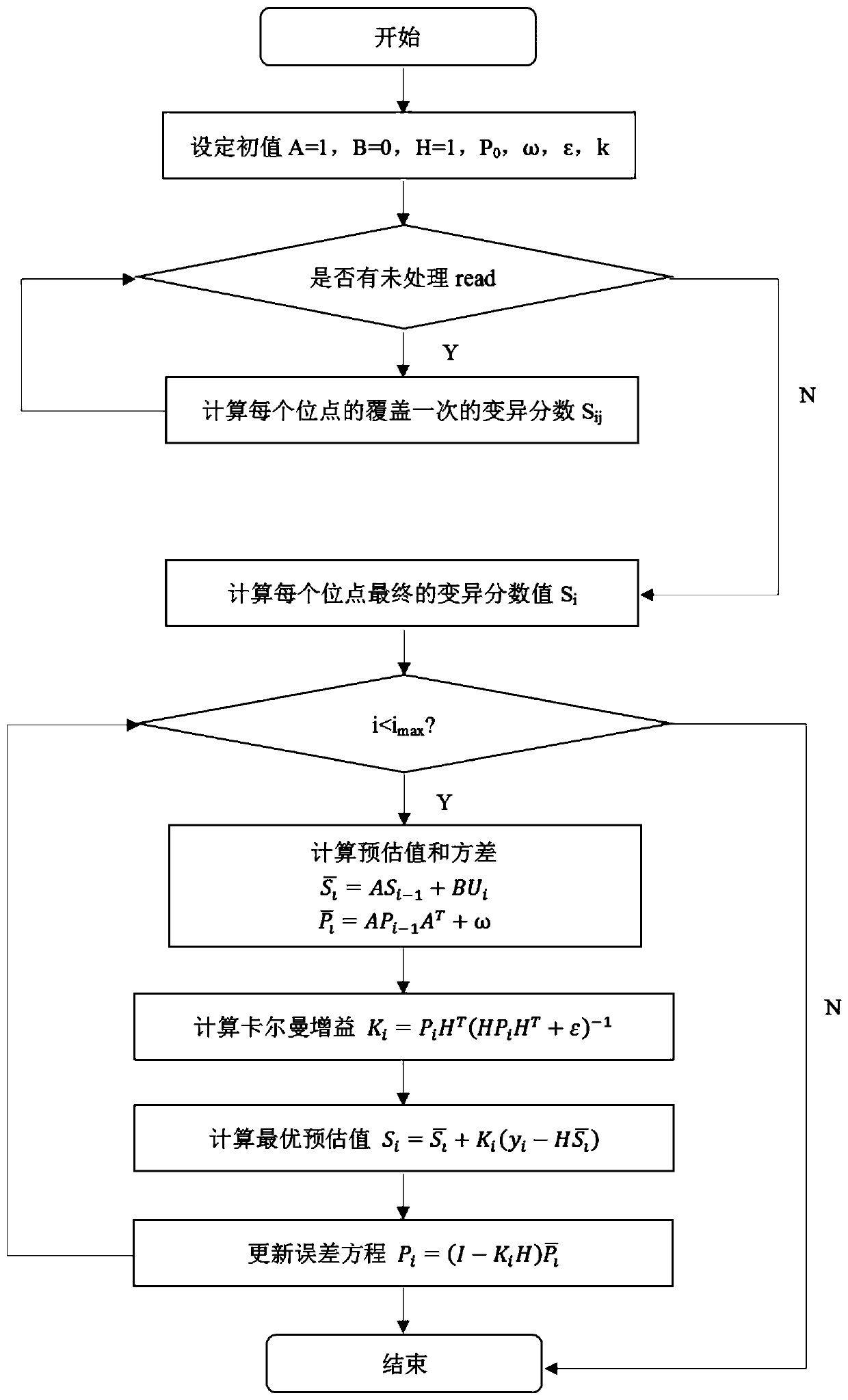
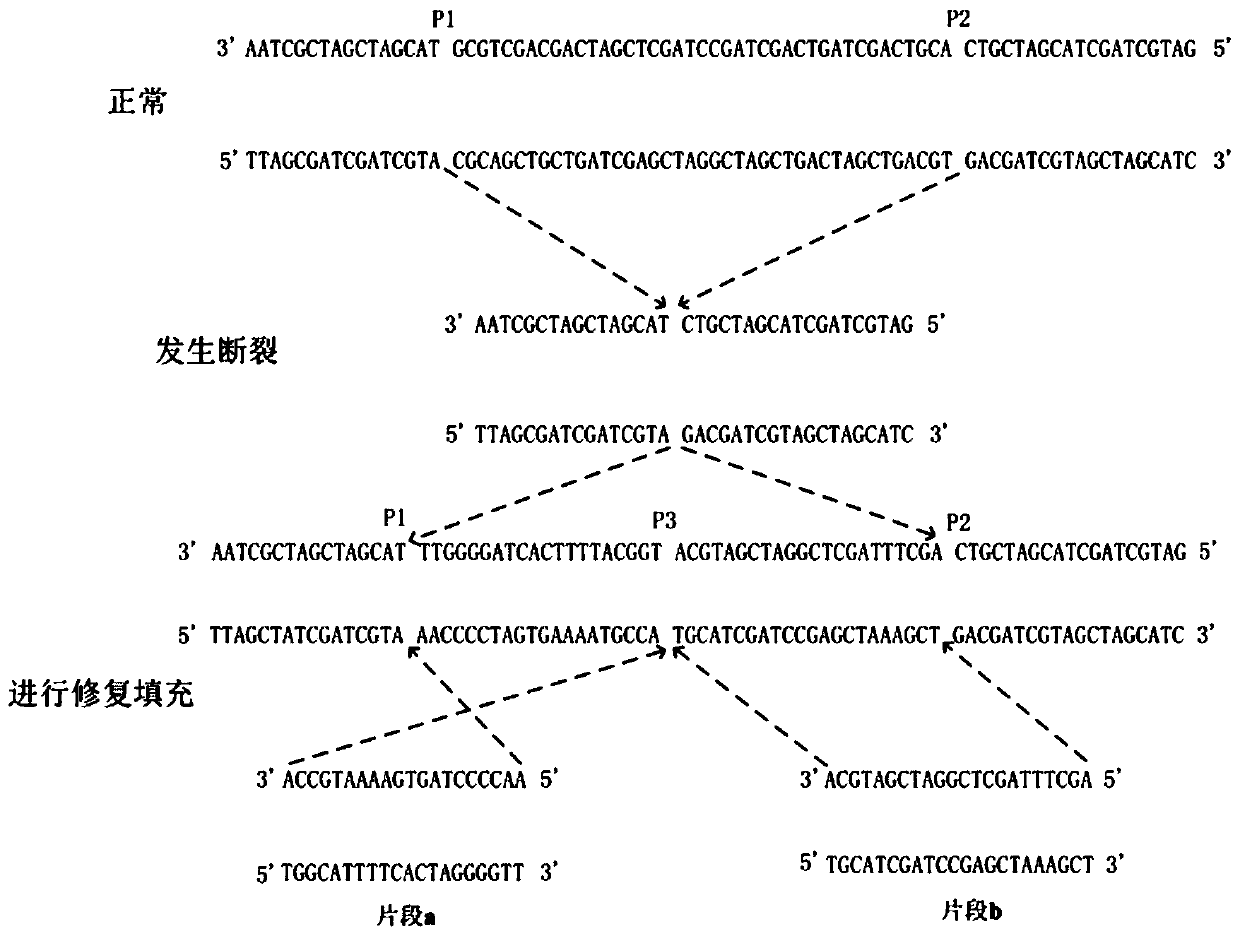
[0080] The present invention provides a highly fault-tolerant genome mutation detection method based on a filtering strategy. A highly fault-tolerant detection algorithm (CIDDⅡ) based on a filtering strategy is a complex indel proposed based on the fault tolerance mechanism of Kalman filter and support vector machine (SVM) Detection algorithm.
[0081] See figure 1 , The present invention is a high-fault-tolerant genome complex structure mutation detection method based on a filtering strategy, including the following steps:
[0082] S1, preprocess the SAM file;
[0083] Traverse all the comparison results in the SAM file. The SAM file has already given the comparison quality of the reads and sorted them accordingly. Each read only processes the results of the best comparison quality, that is, traverses the reads of the best quality comparison. CIGAR field in;
[0084] S2, according to the CIGAR field after the comparison and the variation score calculation criteria, calculate the va...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com