Eclipta prostrate endophytic bacteria, eclipta prostrate composition and application of eclipta prostrate endophytic bacteria and eclipta prostrate composition
A technology of endophytic bacteria and eclipta, applied in the direction of bacteria, microorganisms, microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Screening and Isolation of Endophytic Bacteria in Eclipta
[0033]The surface of fresh Eclipta chinensis was washed with sterile water and dried at room temperature. Under sterile conditions, cut up the roots, stems and leaves respectively and take 100mg in a test tube. Add 1 ml of sterile water and centrifuge the sample at 4000 rpm for 1 min. Each 50 μl of supernatant was spread on LB and MRS plates, and cultured at 37°C for 24 hours. By observing the characteristics of the colonies on the plates, 10 colonies with different phenotypes were screened from the plates for antibacterial tests for further screening.
[0034] Experimental results:
[0035] A total of 10 strains of bacteria were screened from Eclipta, among which, 5 strains were screened from roots, 2 strains from stems and 3 strains from leaves. The 10 strains of bacteria were named EP01-EP10 respectively. Most of the colonies screened from the roots and leaves had common characteristics such ...
Embodiment 2
[0036] Embodiment 2: Molecular biology identification and antibacterial test
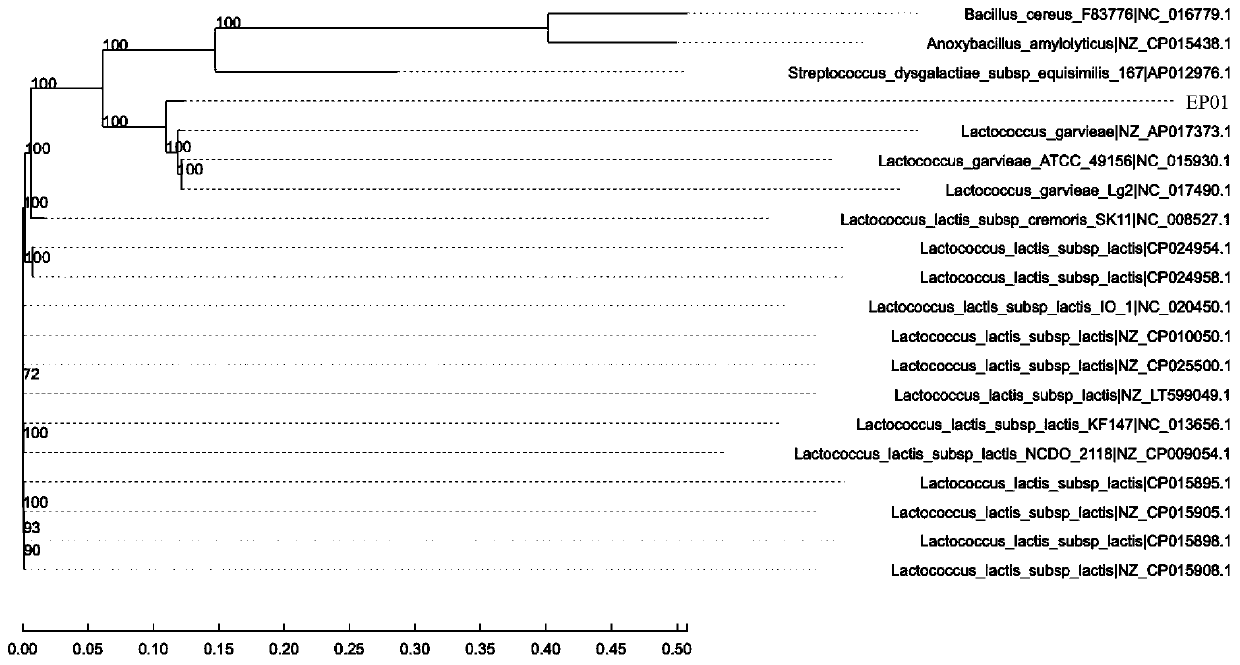
[0037] Genomic DNA of EP01-EP10 strains was extracted by ctab / sds method and used as a template for PCR amplification. The 16s ribosomal DNA sequences of 10 strains were amplified with universal primers 27f: 5'-agagttttgatggctcag-3' (SEQ ID NO: 1) and 1492r: 5'-acggttatccttgttaccgatt-3' (SEQ ID NO: 2) and sequencing. Amplification was performed using polymerase chain reaction in a PCR thermocycler (MyCycler; Bio-Rad Laboratories Inc., USA). The PCR amplification system is 25 μl, including 0.2 μl taq enzyme (0.5u / ml), 2.5 μl 10× buffer, 1.8 μl mg 2+ , 1 μl dntps mix, 1 μl template DNA, 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM) and 17.5 μl ddH 2 O, amplification conditions: 95°C, 3min; 95°C, 30s, 55°C, 60s, 72°C, 90s (30 cycles); 72°C, 5min; 4°C to terminate the reaction. The amplified products were purified and sequenced by Beijing Huada Biotechnology Co., Ltd. According to ...
Embodiment 3
[0046] Example 3: Genome Sequencing and Antimicrobial Drug Mining
[0047] Use Illumina PE150 sequencing technology to sequence the genome of EP01 endophytic bacteria, construct a fragment with an insert fragment of ~400bp from the qualified DNA sample, and perform PE150 (pair-end) sequencing, that is, double-end sequencing, and the read length of single-end sequencing is 150bp , each sample provides the amount of raw sequencing data not lower than the 100× coverage depth of the genome. Use the short sequence assembly software SOAPdenovo2 (http: / / soap.genomics.org.cn / ) to splice the optimized sequence after next-generation sequencing with multiple Kmer parameters to obtain the optimal contigs assembly result, and then compare the reads to On contig, according to the paired-end and overlap relationship of reads, the assembly results are partially assembled and optimized to form scaffolds and obtain the whole genome sequence. (For genomic data see BioProject: PRJNA582067; Bio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com