Multiplex production and barcoding of genetically engineered cells
A genetic engineering, barcoding technology, applied in other directions of inserting foreign genetic material, genetic engineering, biochemical equipment and methods, etc., can solve problems such as limiting the phenotypic selection of characterizing individual variants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
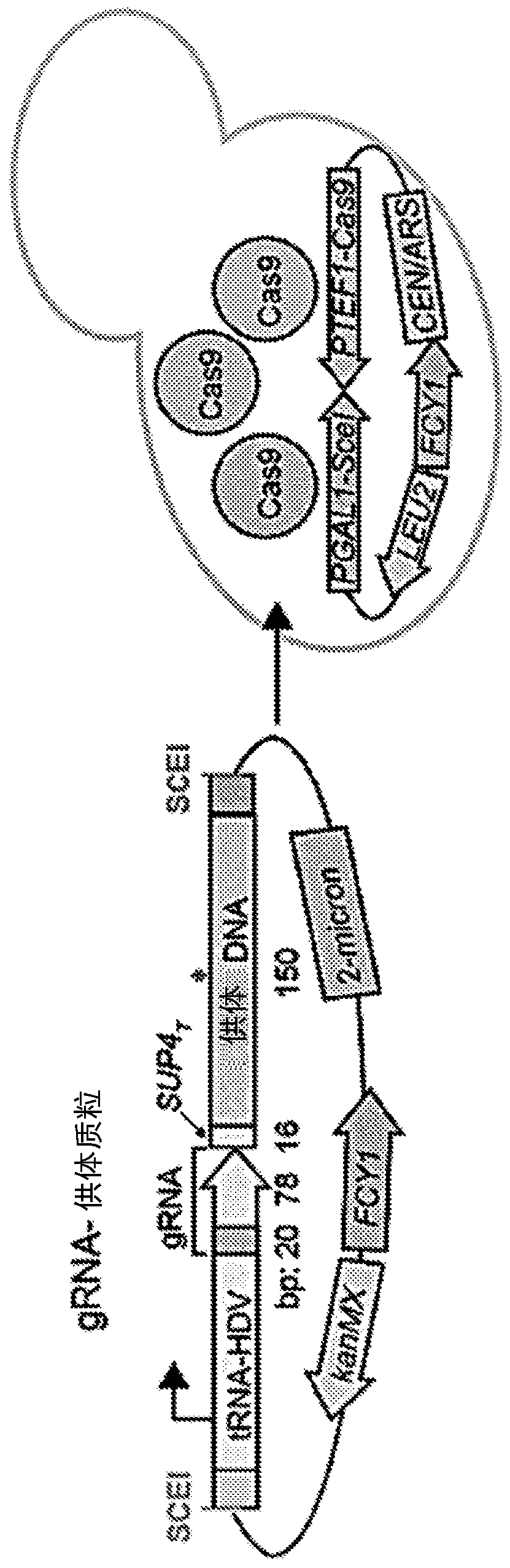
[0200]Example 1 describes the use of the yeast, Saccharomyces cerevisiae, for this purpose. Saccharomyces cerevisiae exists in diploid and haploid forms. Conjugation only occurs between yeast haploid forms of different conjugation types, which can be a or alpha conjugation types. The allele of the MAT locus (MATa or MATα) determines the mating type. Diploid cells were generated from crossing MATa and MATα yeast strains. Thus, haploid genetically modified yeast cells containing a gRNA-donor polynucleotide cassette can be hybridized with haploid barcoded yeast cells to generate diploid yeast cells, including gRNA-donor polynucleotide cassettes on different nucleic acids and barcode sequence. For example, a genetically modified yeast cell of strain MATα can be mated with a barcoded yeast cell of strain MATa. Alternatively, genetically modified yeast cells of strain MATa can be mated with barcoded yeast cells of strain MATa.
[0201] The gRNA-donor polynucleotide cassette is ...
Embodiment 2
[0341] Example 2. Genome Editing Using the Cpf1-Donor System Produces Efficient Editing
[0342] When the Cpf1 guide-donor system was used in a similar manner as described in Example 1, the Cpf1 guide-donor system caused efficient (>99%) editing and ~10-fold improvement with Cpf1 editing, with a degree of donor recruitment similar to Cas9.
[0343] Figure 14A and 14B provide data. Figure 14A Cell colonies showing pre-expression of Cpf1 were transformed with a Cpf1 guide-donor plasmid targeting the ADE2 gene (the guide has a Cpf1 scaffold). Donor DNA encodes a mutation that causes a frameshift. Figure 14B Shows % red colonies (ratio of red:white colonies) when Cpf1 guide-donor and non-editing plasmids were mixed at a ratio of 17:3 and transformed into Cpf1-expressing cells, without (left) or with (right) LexA-FHA .
Embodiment 3
[0344] Example 3. Plasmid Spike-In experiment proves that LexA-FHA and linearized vector improve HDR efficiency and editing survival.
[0345] ADE2ORF-editing plasmids were mixed at 85% (17:3) with non-editing plasmids and transformed into Cas9-carrying ( Figure 16 , top panel) or Cas9 and LexA-FHA ( Figure 16 , bottom panels) strains. Using the same strain for each transformation made direct comparison of total colonies per row feasible.
[0346] Figure 16 provide data. The y-axis indicates the total number of colonies observed in each transformation, while the x-axis indicates the percentage of colonies in red, which represents the survival of the process of editing ADE2. The shape of each spot corresponds to the restriction enzyme used to linearize the plasmid outside the transformation precursor. Five different columns correspond to different forms of spike-in mixtures. The first number corresponds to the number of genomic loci cleaved by the ADE2 editing plasmid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com