NGS-based chromosome equilibrium translocation detection and analysis system and application
A balanced translocation and analysis system technology, applied in the field of chromosome balanced translocation detection and analysis system, can solve the problems of high false positive rate, excessive time consumption, complicated process, etc., and achieve wide application range, high analysis accuracy and simple steps Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
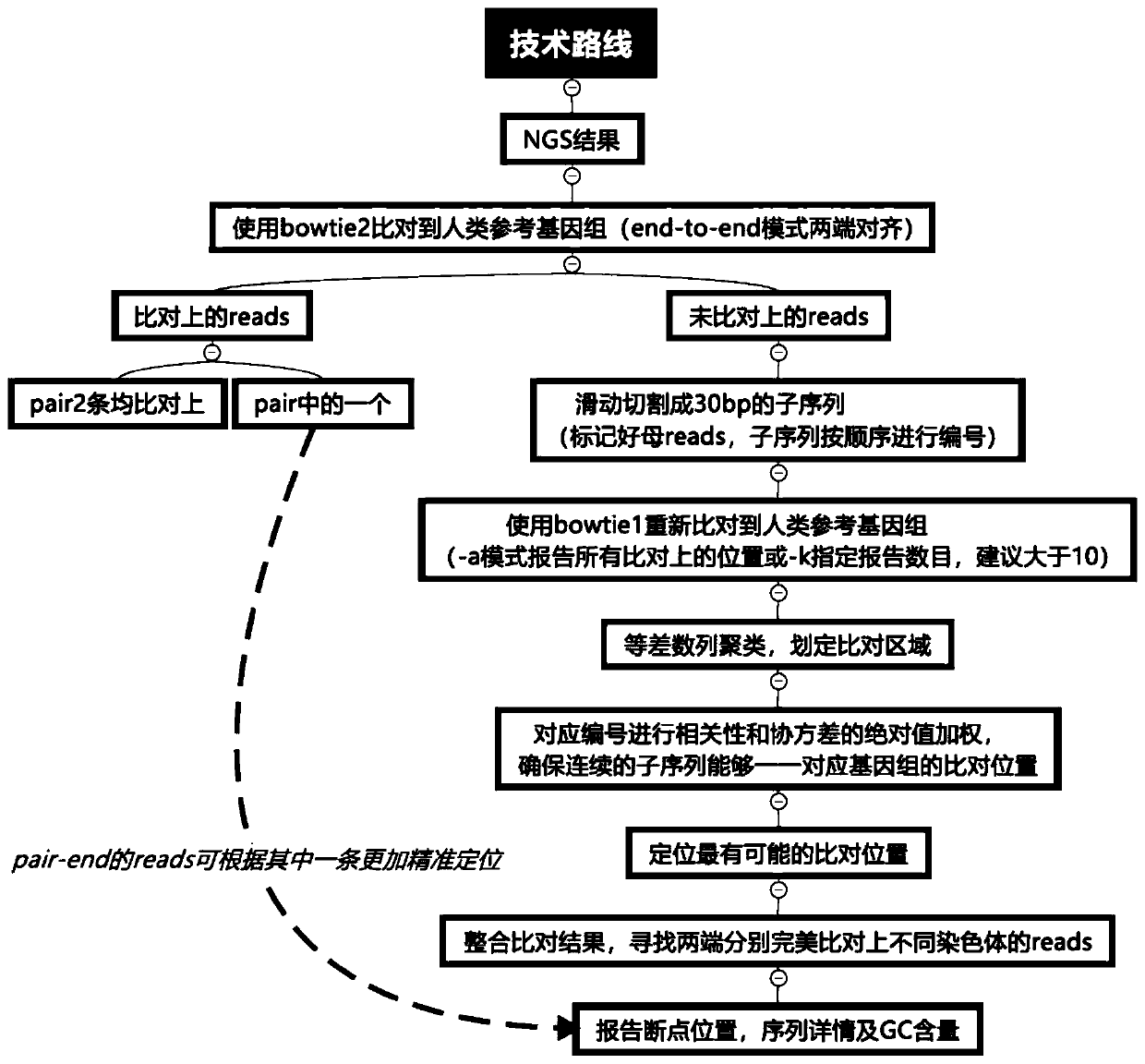
[0043] A method for detecting and analyzing chromosome balanced translocation based on NGS is carried out according to the following steps, and its technical route is as follows: figure 1 shown.
[0044] 1. Data acquisition.
[0045] Obtain the reads data obtained by NGS detection. In order to ensure that the fastq sequences used for comparison meet the quality requirements, use the fastq data quality control filtering software and fastp software to filter the fastq files obtained from the sequencing result data.
[0046] 2. Genome comparison.
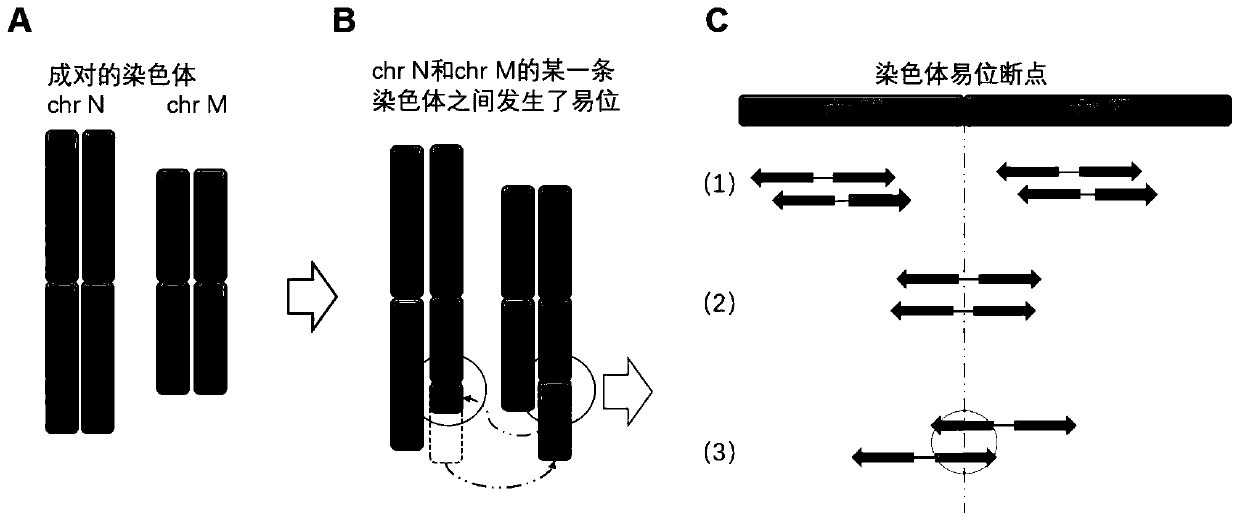
[0047] figure 2 It is a schematic diagram of a balanced translocation of chromosomes; where: A indicates a paired chromosome, B indicates a translocation between chromosomes, and C indicates that there are three situations of reads obtained by sequencing after the translocation occurs.
[0048] (1) The paired reads can be completely aligned to the chromosome chr M or chr N of the human genome.
[0049] (2) One of the paired reads ...
Embodiment 2
[0282]An NGS-based balanced chromosome translocation detection and analysis system, including: a data acquisition module, a genome comparison module, a sliding cutting module, a short sequence comparison module, a breakpoint analysis module and a result output module.
[0283] The data acquisition module is used to acquire the reads data detected by NGS;
[0284] The genome comparison module is used to compare the above-mentioned reads with the human reference genome respectively, and obtain the first set of reads that cannot match the human reference genome;
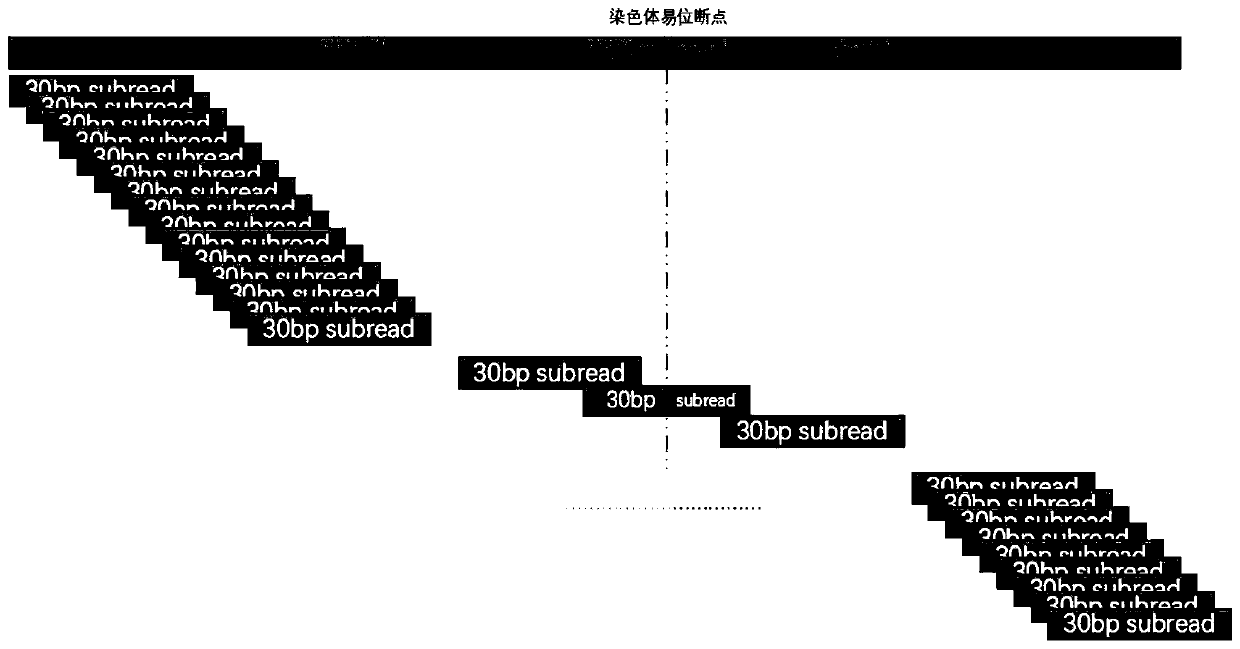
[0285] The sliding cutting module is used to perform sliding cutting on the reads in the first reads set to obtain the subsequences of each reads, and use the arithmetic sequence to mark and number to obtain the second reads set composed of the above subsequences;
[0286] The short sequence alignment module is used to compare each subsequence in the above-mentioned second reads set with the human reference genome, and ...
Embodiment 3
[0291] Using the NGS-based balanced chromosome translocation detection and analysis system of Example 2, the sample data of two cases were analyzed, and after the reads data obtained by NGS detection were obtained, genome alignment, sliding cutting, and short sequence alignment were performed to obtain the following The two ends are respectively compared to the third reads set of different chromosomes of the human reference genome.
[0292] Table 1. The third reads set obtained from the analysis of two samples
[0293]
[0294] The above-mentioned reads were analyzed and calculated, and the result of balanced chromosome translocation was obtained as follows.
[0295] Table 2. Balanced Chromosomal Translocations
[0296]
[0297]
[0298] Note: In the above table of matching chromosome position results, the last 1 or -1 indicates the correlation analysis results, all of which are 1 or -1.
[0299] The above-mentioned samples were simultaneously tested for balanced ch...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com