Method for knocking in terminator to achieve transcription factor knockout by using gene editing technology
A technology of gene editing and transcription elements, applied in the biological field, can solve problems such as difficulty in picking out single clones, experimental deviations, and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
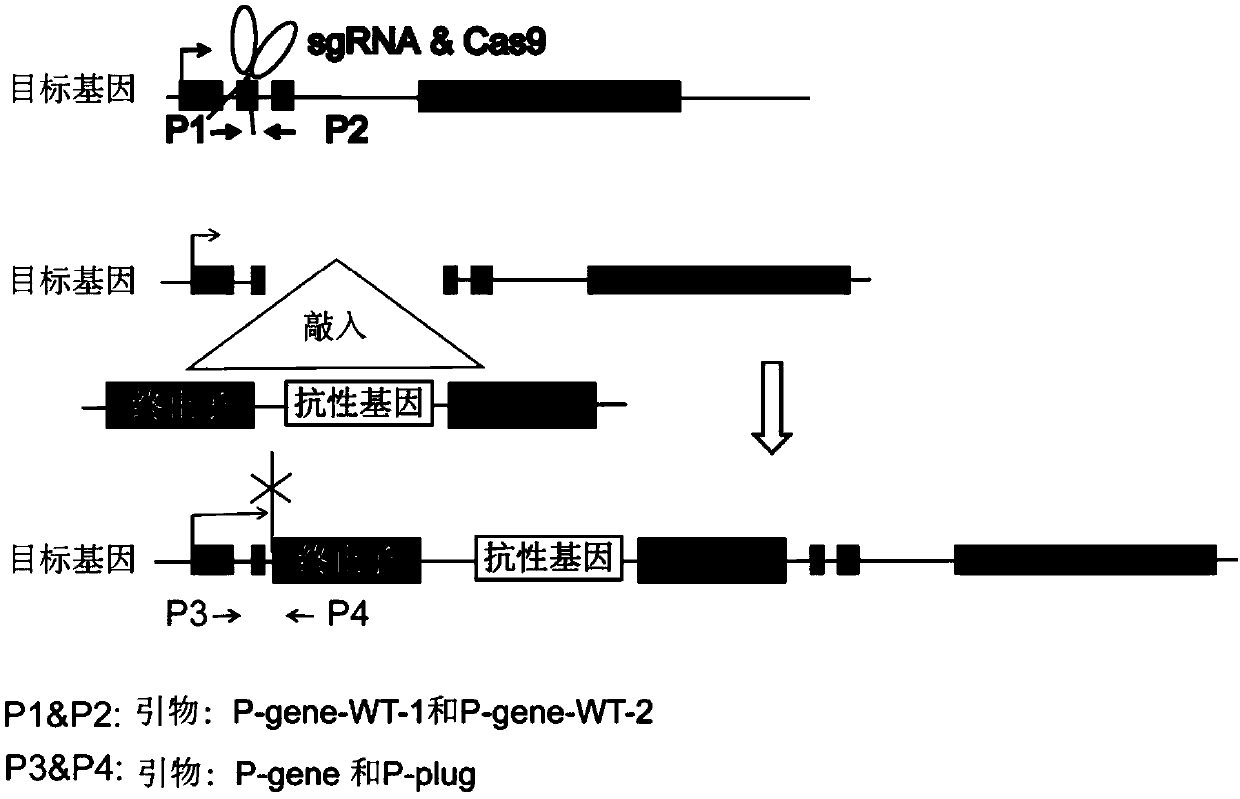
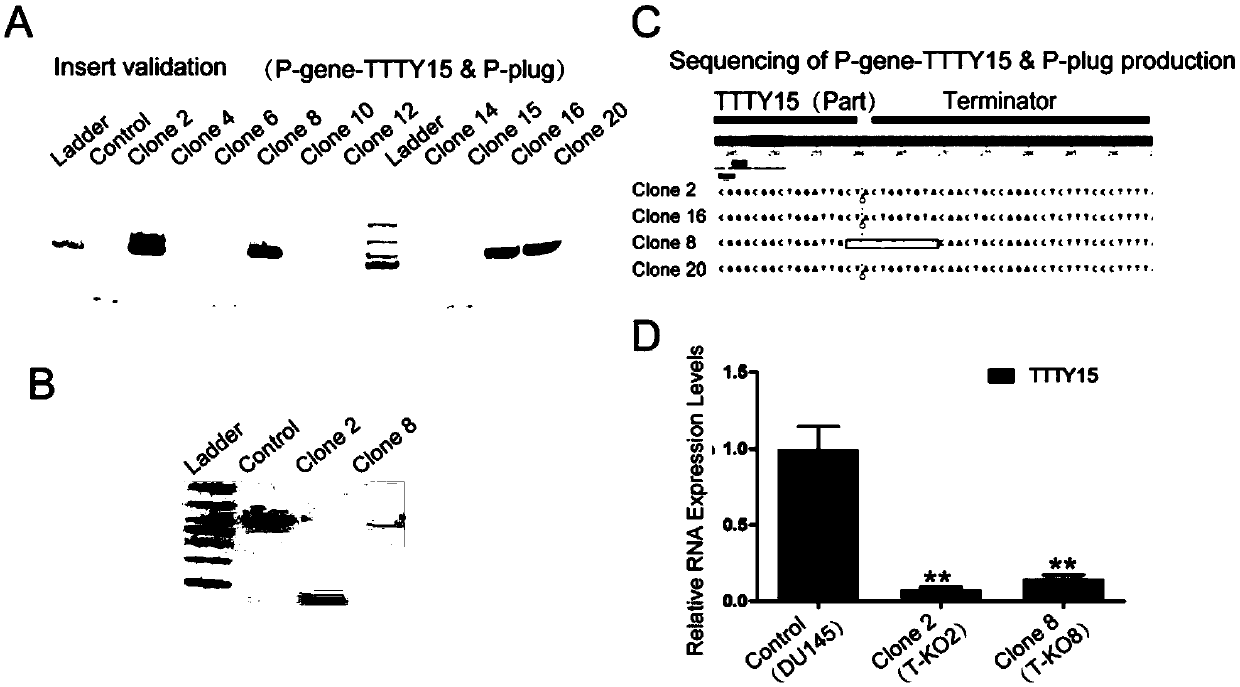
[0113] Example 1: CRISPR-Cas9 technology knocks in terminator to realize knockout of long non-coding RNA TTTY15
[0114] 1) Using molecular cloning technology to construct a vector containing the transcription termination sequence (terminator) TK_PA_terminator and containing the Neomycin resistance gene, and expressing EGFP.
[0115] The specific sequence of TK_PA_terminator is shown in SEQ ID NO.1.
[0116] 2) The human genome in mammals was selected as the design intervention target, TTTY15 was used as the non-coding RNA to be knocked out, and the gRNA was designed to recognize: gRNA-TTTY15, and its recognition site sequence is as follows, TCAGGCAATCCAGCCGCCC GGG (SEQ ID NO.2), wherein GGG is a PAM sequence.
[0117] 3) The gRNA-TTTY15 and gRNA-plug expression vectors were constructed respectively, and the corresponding expression elements were constructed into the lentiviral backbone vector PLKO-bsd (these two plasmids can be packaged into lentiviruses). The Cas9 express...
Embodiment 2
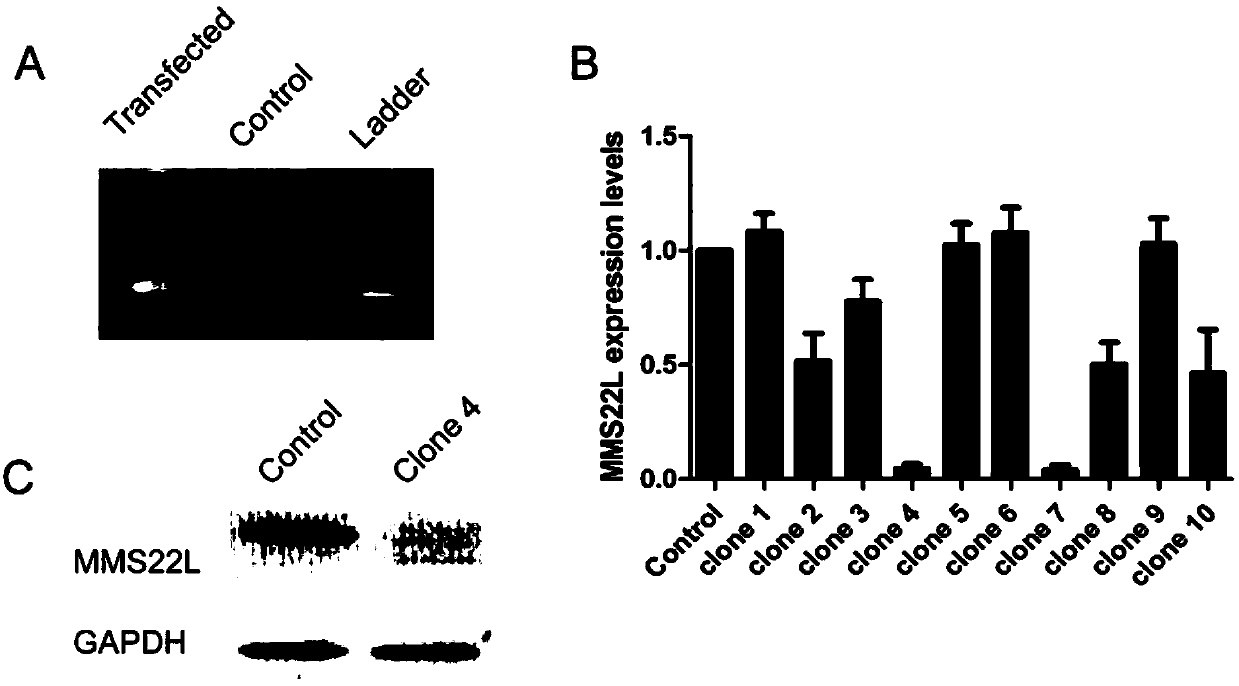
[0137] Example 2: CRISPR-Cas9 technology knocks in terminator to realize knockout of protein coding gene MMS22L
[0138] 1) Using molecular cloning technology to construct a vector containing the transcription termination sequence (terminator) TK_PA_terminator and containing the Neomycin resistance gene, and expressing EGFP.
[0139] 2) The human genome in mammals was selected as the design intervention target, MMS22L was used as the protein-coding gene to be knocked out, and the gRNA was designed to recognize: gRNA-MMS22L, and its recognition site sequence is as follows, GCTTAAGGGCTCCGCTGCAG AGG (SEQ ID NO.3), wherein AGG is a PAM sequence.
[0140] 3) The expression vectors of gRNA-MMS22L and gRNA-plug were respectively constructed, and the corresponding expression elements were constructed into the lentiviral backbone vector PLKO-bsd (these two plasmids can be packaged into lentiviruses). The Cas9 expression vector is a commercially available plasmid Cas9 expression vector ...
Embodiment 3
[0151] Example 3: CRISPR-Cas9 technology knocks in the terminator to realize the knockout of microRNA miR-155
[0152] 1) A vector containing the transcription termination sequence (terminator) TK_PA_terminator (flanked with TTTT sequence) and the Hygromycin B resistance gene was constructed using molecular cloning technology.
[0153] 2) The human genome in mammals is selected as the design intervention target, miR-155 is used as the microRNA to be knocked out, and the gRNA is designed to recognize: gRNA-miR-155, and its recognition site sequence is as follows, GTTAATGCTA ATCGTGATAG GGG (SEQ ID NO.4), wherein GGG is a PAM sequence.
[0154] 3) The expression vectors of gRNA-miR-155 and gRNA-plug were respectively constructed, and the corresponding expression elements were constructed into the lentiviral backbone vector PLKO-bsd (these two plasmids can be packaged into lentivirus). The Cas9 expression vector is a commercially available plasmid Cas9 expression vector was purc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com