Protein-protein interaction prediction method based on deep forest
A prediction method and protein technology, applied in the field of biological information, can solve problems such as high cost, and achieve the effect of improving prediction accuracy and reducing model complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
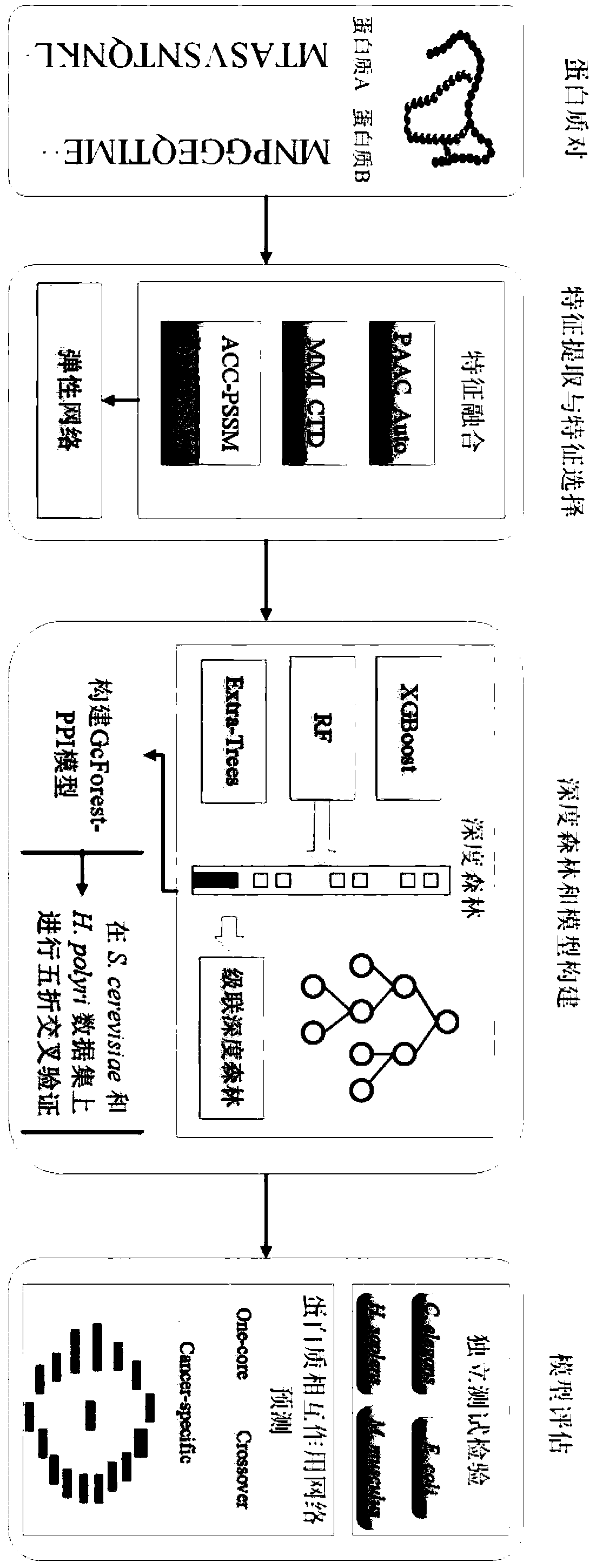
[0082] A deep forest-based protein-protein interaction prediction method GcForest-PPI, the specific steps are as follows (such as figure 1 shown):
[0083] 1) collect data
[0084] 1-1) Select the protein interaction data set of yeast S.cerevisiae and Helicobacter pylori H.pylori as the training set, and use the experimentally verified protein-protein interaction pairs obtained from the database as positive samples. Interacting protein pairs serve as negative samples. The S.cerevisiae dataset is from the DIP (www.dip.doe-mbi.ucla.edu) core database, version DIP_20070219. H. pylori is derived from Martin, S., Roe, D. and Faulon, J.L. (2005). Predicting protein-protein interactions using signature products. Bioinformatics, 21(2), 218-226.
[0085] 1-2) For the yeast data set, first remove samples with less than 50 residues in the sample, and then use the multiple sequence alignment tool CD-HIT program to remove protein sequences with sequence similarity higher than 40%, and o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com