Enzymes for glycan analysis
A technology for glycosidase and glycoprotein, which is applied in the field of tools for glycan analysis, and can solve the problems of difficulty, reduction, and function of glycoprotein research.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0047] Polypeptides can be produced by suitable methods, including recombinant or synthetic methods. For example, polypeptides can be synthesized directly using standard techniques known in the art, such as Fmoc solid phase chemistry, Boc solid phase chemistry, or by solution phase peptide synthesis. Alternatively, a polypeptide may be produced by transforming a cell, usually a bacterial cell, with a nucleic acid molecule or vector encoding the polypeptide. Methods for producing polypeptides by expression in bacterial host cells are described below and exemplified in the Examples. The invention provides nucleic acid molecules and vectors encoding polypeptides of the invention. The invention also provides host cells comprising such nucleic acids or vectors. An exemplary polynucleotide molecule encoding a polypeptide disclosed herein is provided as SEQ ID NO:13. The sequence includes codons for an N-terminal methionine (ATG) at the 3' end and codons for a GSGLE linker and a 6...
Embodiment 1
[0128] Example 1 - Sialidase
[0129] Materials and Methods
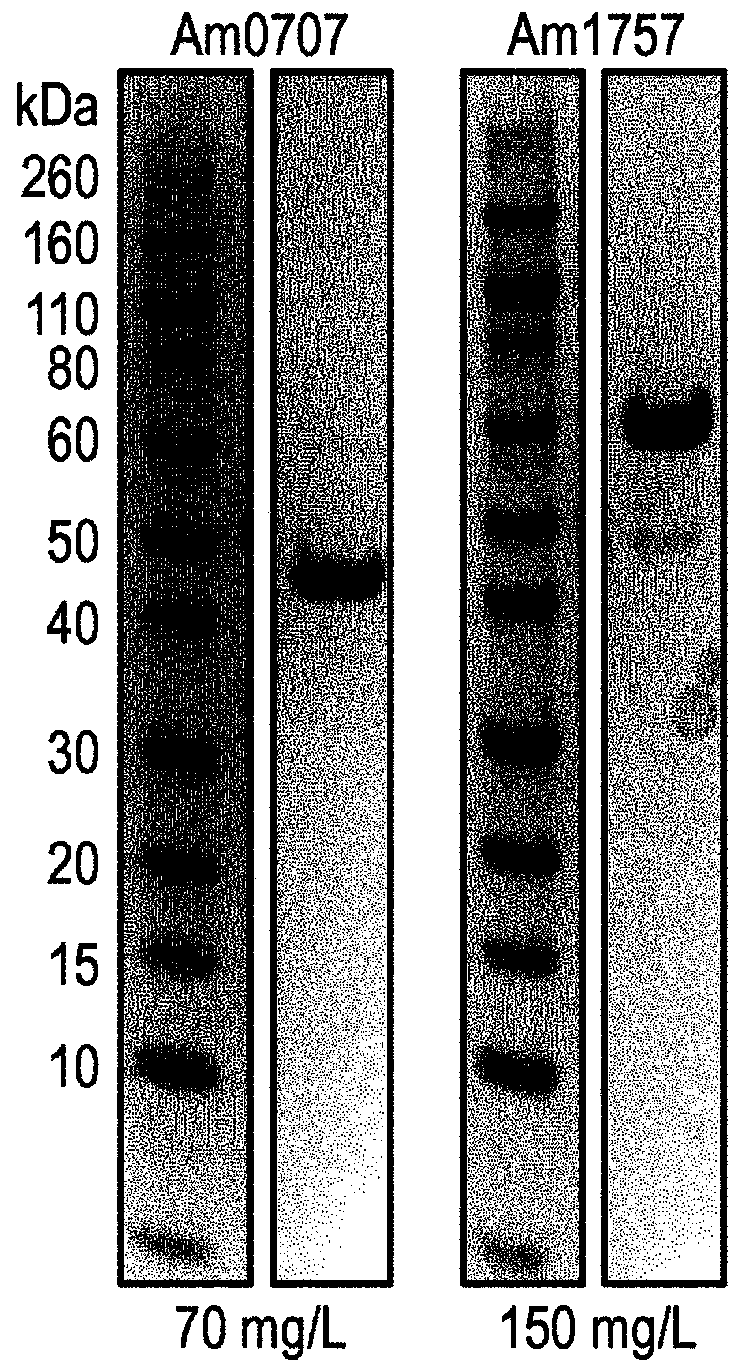
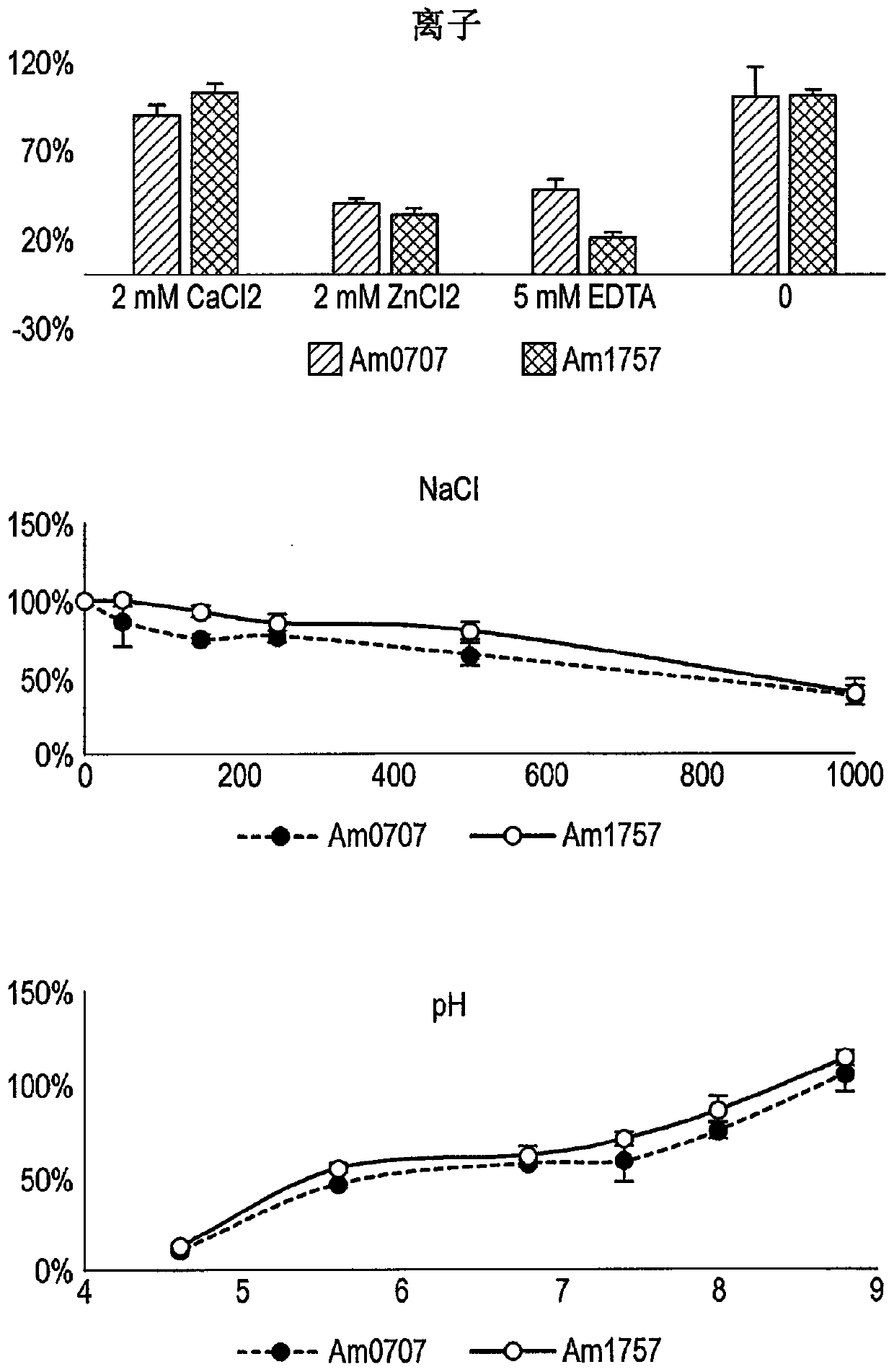
[0130] Expression and purification of sialidase
[0131] The genes identified in Ekkermansia muciniphila (Am0705, Am0707, Am1757, Am2058) were codon optimized for good expression in E. coli in the vector pET21a(+). This vector was transformed into BL21(DE3)Star cells. E. coli was routinely grown in LB at 37°C, 200 rpm. In the presence of the plasmid, add 100 µg / mL kanamycin. After overnight incubation, dilute the culture 1:20 in fresh LB(amp) and grow the culture until OD 620 About 0.7 to 0.8, after which recombinant protein expression was induced by adding 1 mM IPTG, and the expression lasted for 5 hours, and then the cells were collected and frozen. Frozen cells were thawed and dissolved in His-binding buffer (20 mM NaP pH 7.4, 500 mM NaCl, 20 mM imidazole) and sonicated to release intracellular proteins. Cell debris was removed by centrifugation. The filtered sterile supernatant was affinity purified on ...
Embodiment 2-O
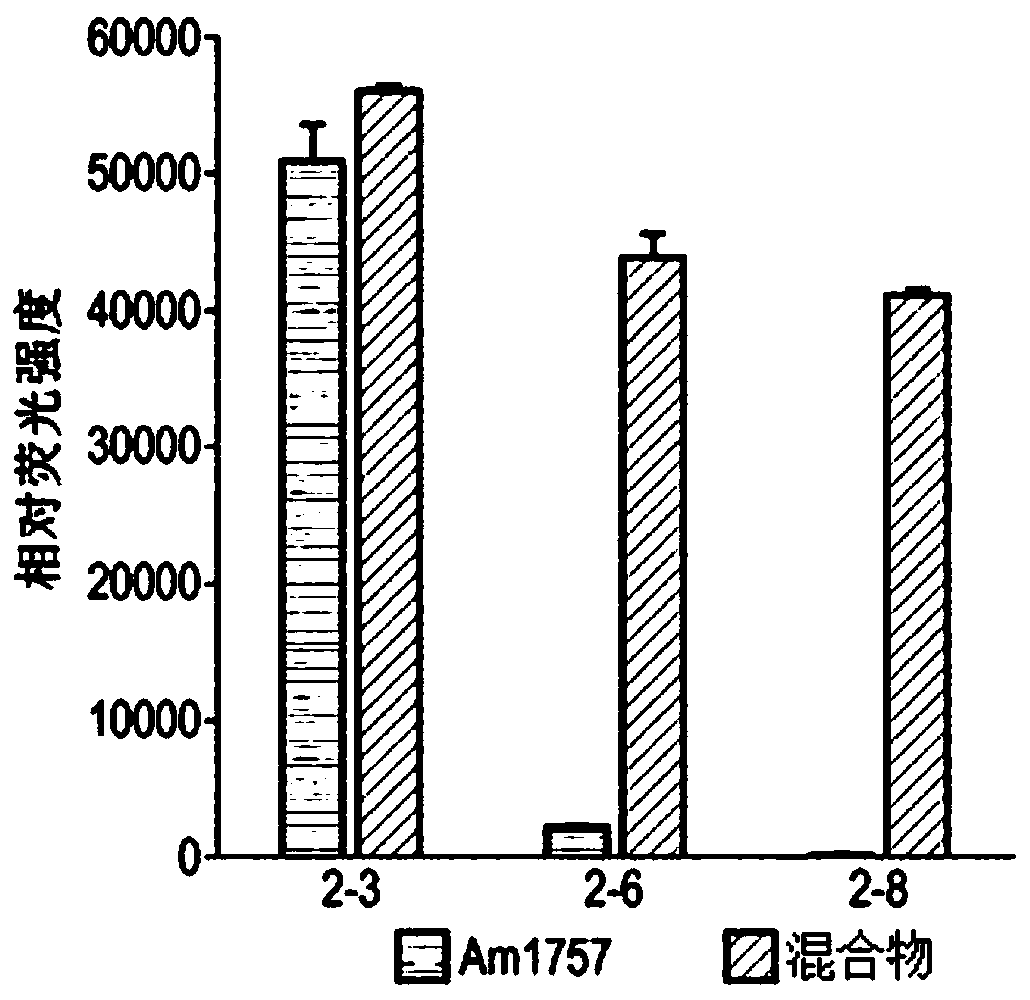
[0159] Example 2-O-glycosidase
[0160] Materials and methods
[0161] Expression and purification of endo-O-glycosidase
[0162] The endo-N-acetylgalactosaminidase of Streptococcus oralis was codon optimized for good expression in E. coli in the vector pET21a(+). This vector was transformed into BL21(DE3)Star cells. E. coli was routinely grown in LB at 37°C, 200 rpm. In the presence of the plasmid, add 100 µg / mL kanamycin. After overnight incubation, dilute the culture 1:20 in fresh LB(amp) and grow the culture until OD 620 About 0.7 to 0.8, after which recombinant protein expression was induced by adding 1 mM IPTG, and the expression lasted for 5 hours, and then the cells were collected and frozen. Frozen cells were thawed and dissolved in His-binding buffer (20 mM NaP pH 7.4, 500 mM NaCl, 20 mM imidazole) and sonicated to release intracellular proteins. Cell debris was removed by centrifugation. Filtered sterile supernatants were affinity purified on a nickel colum...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com