Method for accurately determining bacteria in special condition on basis of microfluidic visualization technology as well as selecting and enriching bacteria in special condition
A special state and microfluidic technology, applied in the direction of microorganism-based methods, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem that the number of bacteria in special states cannot be accurately quantified, and achieve high efficiency and high dosage. Less, precise process control effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The specific operation steps for obtaining a single droplet encapsulated single cell in the present invention are as follows:
[0037] 1. Streak the bacteria stored in the -80°C glycerol tube on the LB plate, and place it at 37°C for constant temperature incubation for 18-24 hours. Pick a single colony on the plate, inoculate it in LB broth, and cultivate at 37°C for 12h.
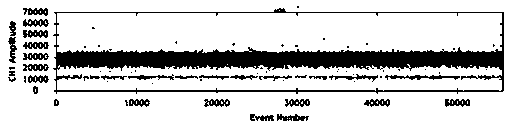
[0038] The bacterial solution was diluted with sterile physiological saline, and the appropriate gradient bacterial solution was selected as the water phase. Microfluidic devices were used to form microdroplets, and a digital PCR instrument was used to measure the copy number of the initial sample. like figure 1 As shown, the measured initial bacterial concentration was 5.4545×10 4 CFU / mL. Take 880 μL of the bacterial solution and resuspend it in 1 mL of normal saline to obtain a concentration of 4.8×10 4 CFU / mL sample solution, and resuspended in LB broth, so that the number of bacteria in the s...
Embodiment 2
[0043] Taking Cronobacter sakazakii in VBNC state as an example - counting, sorting and enrichment of bacteria in special state:
[0044] 1. Streak the bacteria stored in the -80°C glycerol tube on the LB plate, and place it at 37°C for constant temperature incubation for 18-24 hours. Pick a single colony on the plate, inoculate it in LB broth, and cultivate at 37°C for 6-7h.
[0045] 2. Take 30mL logarithmic phase bacterial solution (about 10 8 CFU / mL) was centrifuged, and the pellet was transferred to 300 mL of sterile normal saline, and 120 mg of ampicillin was added to the normal saline for induction at room temperature.
[0046] 3. Take 1 mL of the bacterial solution during the induction process, add 5 μL PMAxx dye, and incubate it in the dark for 15 min, then expose it in a nucleic acid exposure device for 15 min, and use the DNA extraction kit to extract DNA for digital PCR detection. Use the copy number to determine the bacterial concentration of the viable bacterial...
Embodiment 3
[0054] Take Staphylococcus aureus as an example—screening for antibiotic-resistant strains.
[0055] 1. Streak the golden yellow grape balls stored in a glycerol tube at -80°C on the LB plate, and place them at 37°C for incubation for 18-24 hours. Pick a single colony on the plate, inoculate it in LB broth, and cultivate at 37°C for 12h.
[0056] 2. Gradient dilution of the bacterial solution with sterile normal saline, use digital PCR to determine the bacterial concentration in the sample, and adjust the bacterial concentration to 4.8×10 according to the copy number. 4 CFU / mL.
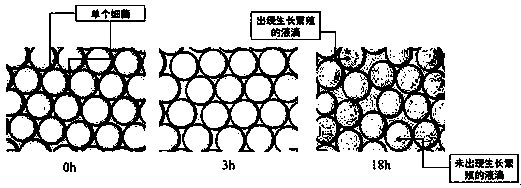
[0057] 4, will be 4.8 × 10 4 The bacterial liquid of CFU / mL was resuspended in LB liquid medium containing penicillin as the water phase to generate droplets, which were transferred to an eight-connected tube and incubated at 37°C for 18h at a constant temperature, which can be resistant to antibiotics. The Staphylococcus aureus can reproduce during the culture process.
[0058] 5. After 18 hours ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com