Primer sequence and kit for detecting glucose-6-phosphate dehydrogenase (G6PD) gene mutation
A primer sequence, mutation detection technology, applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of long detection process, technical difficulty, open system, etc. The result is easy to analyze and the effect of improving the work level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
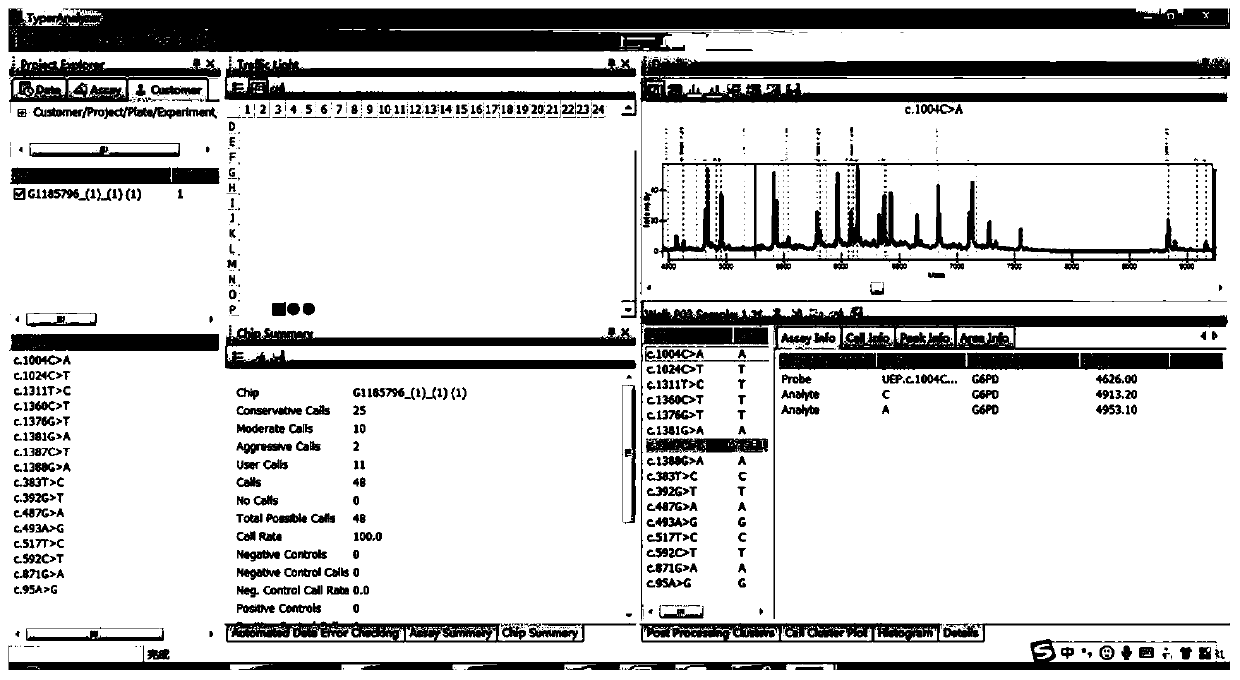
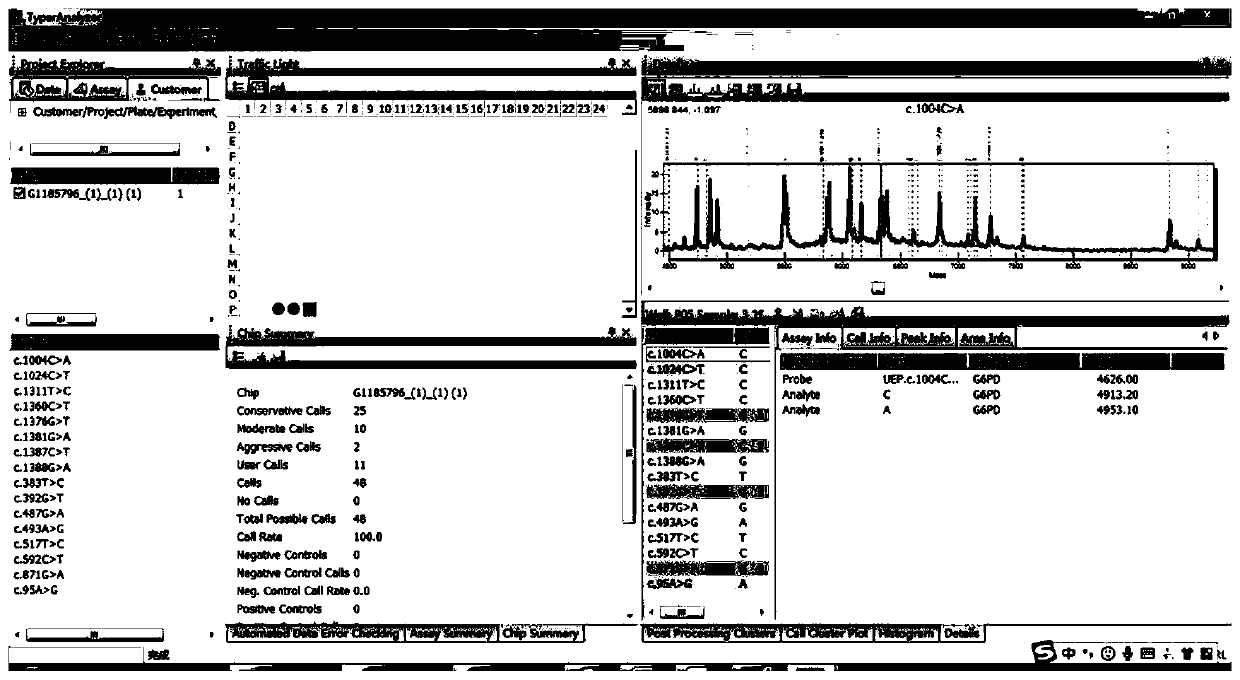
Image
Examples
Embodiment 1
[0043] Example 1: The primers for detecting G6PD gene mutations were synthesized by Shanghai Bailige Biotechnology Co., Ltd., and the sequence is as follows: Amplification primers:
[0044]
[0045] Single base extension primers:
[0046]
[0047]
Embodiment 2
[0048] Example 2: Mutant plasmids for detection of common pathogenic mutations in the G6PD gene were synthesized by Shanghai Sangon Bioengineering Co., Ltd., a total of 3 mutant plasmids (pUC57 clone plasmid + insert sequence), covering more than 99% of the common G6PDd pathogenic mutations in the Chinese population. The incidence of disease mutations is as follows:
[0049]
[0050] Note: *The SNP number, common name and mutation frequency data of the corresponding mutation sites come from the global shared SNP database NCBI dbSNP (https: / / www.ncbi.nlm.nih.gov / snp / ), OMIM database (http: / / www.omim.org / ), the International 1000 Genomes SNP Database (https: / / www.ncbi.nlm.nih.gov / variation / tools / 1000genomes / ), literature, and relevant domestic guidelines and reports in China.
Embodiment 3
[0051] Example 3: Preparation of a detection kit for common pathogenic mutations in the G6PD gene.
[0052] (1) Time-of-flight mass spectrometry detection system nucleic acid sample pretreatment reagent, including the following main components:
[0053]
[0054] (2) Amplification reaction primer premix: a mixture of the nucleotide sequences shown in SEQ ID NO: 1-10; wherein the concentration of primers shown in SEQ ID NO: 1-6 is 0.5 μM, and SEQ ID NO: The concentration of the primers shown in 7-8 is 1 μM, and the concentration of the primers shown in SEQ ID NO: 9-10 is 2 μM;
[0055] (3) Single-base extension reaction primer premix: a mixture of the nucleotide sequences shown in SEQ ID NO: 11-26 in claim 2, the molar concentration of each extension primer is as follows: SEQ ID No.11 : SEQ ID No.12: SEQ ID No.13: SEQ ID No.14: SEQ ID No.15: SEQ ID No.16: SEQ ID No.17: SEQ ID No.18: SEQ ID No.19: SEQ ID No. 20: SEQ ID No.21: SEQ ID No.22: SEQ ID No.23: SEQ ID No.24: SEQ ID ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com