High resolution melting (HRM) typing primer for detecting rs 909253 locus and application of HRM typing primer
A technology for typing primers and loci, applied in the field of molecular biology, can solve problems such as difficult identification of homozygous sequence mutations, and achieve the effects of low cost, good specificity and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: HRM typing primers for detecting rs909253 locus
[0029] The present invention provides an HRM typing primer composition for detecting the rs909253 site, which consists of primer F1 and primer R1:
[0030] Preferably, the nucleotide sequence of the primer F1 is shown in SEQ ID NO.1; the nucleotide sequence of the primer R1 is shown in SEQ ID NO.2.
[0031] The primer F1 and the primer R1 are used for detecting the single nucleotide polymorphism at the rs909253 site by HRM typing.
[0032] At the same time, the present invention also provides an HRM typing reagent composition for detecting the rs909253 site;
[0033] Described HRM typing reagent is made up of primer, dNTPs, qRT-PCR amplification buffer and DNA polymerase;
[0034] The primers are composed of primer F1 and primer R1;
[0035] The invention provides an HRM typing reagent composition for detecting rs909253 site, the dNTPs are composed of fluorescein-labeled dGTP, fluorescein-labeled dATP, fluo...
Embodiment 2
[0039] Example 2: HRM typing primers for detecting rs909253 site
[0040] 1. Extraction of Genomic DNA from Whole Blood
[0041] Collect 1 mL of peripheral blood from the cubital vein of each subject, and use the DP304 kit produced by Tiangen Biochemical Technology (Beijing) Co., Ltd. for DNA extraction.
[0042] (1) Draw 300 μL of fresh blood into a 1.5mL centrifuge tube, add 900 μL of erythrocyte lysate, invert and mix well, place at room temperature for 5 minutes, during which time invert and mix several times, centrifuge at 10000rpm for 1min, absorb the supernatant, leave the white blood cell precipitate, add 200 μL buffer GA, shake until thoroughly mixed.
[0043] (2) Add 20 μL of Proteinase K solution and mix well.
[0044] (3) Add 200 μL buffer GB, mix thoroughly by inversion, place at 70°C for 10 minutes, the solution should become clear, and briefly centrifuge to remove water droplets on the inner wall of the tube cap.
[0045] (4) Add 300 μL of absolute ethanol, s...
Embodiment 3
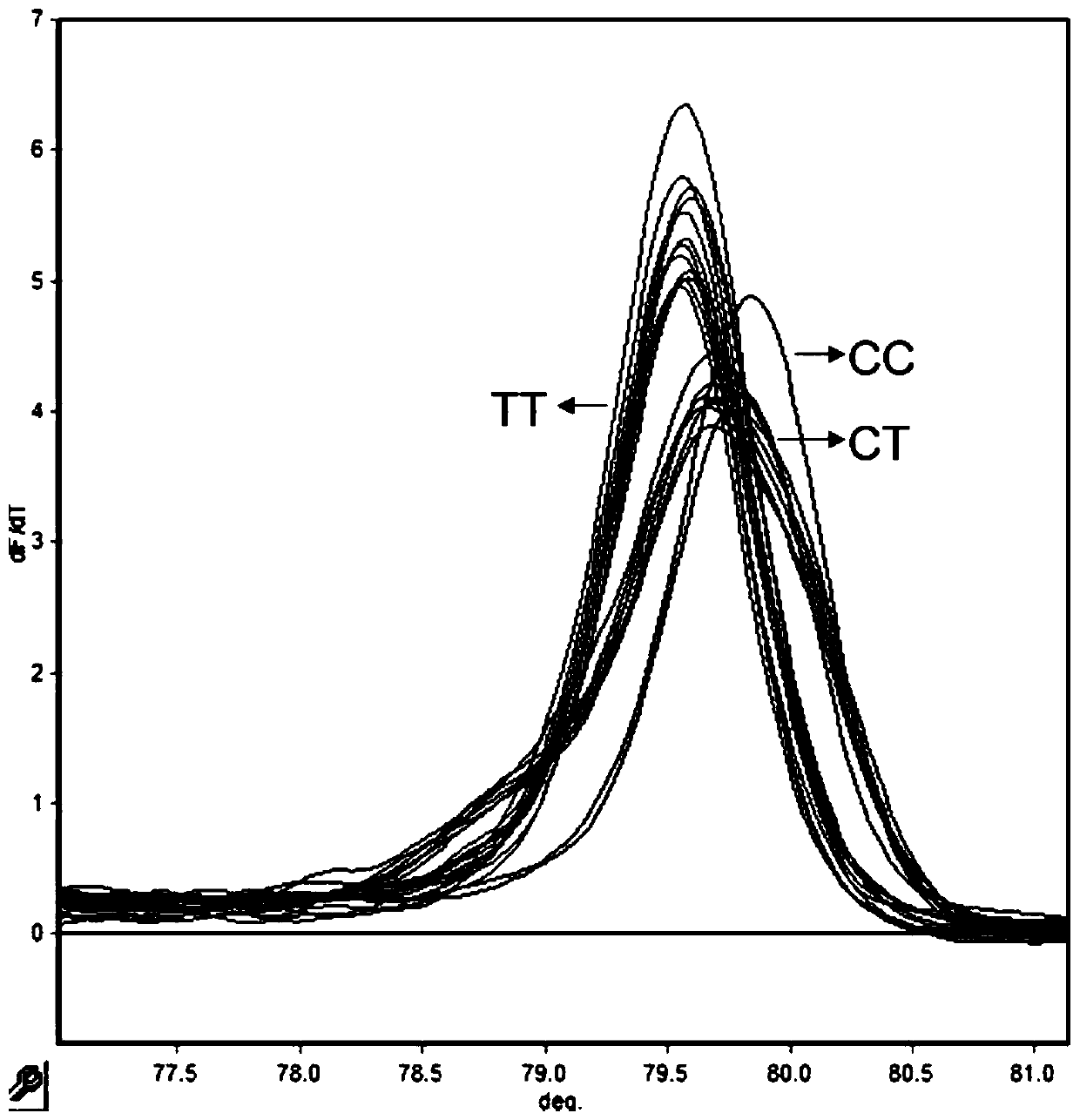
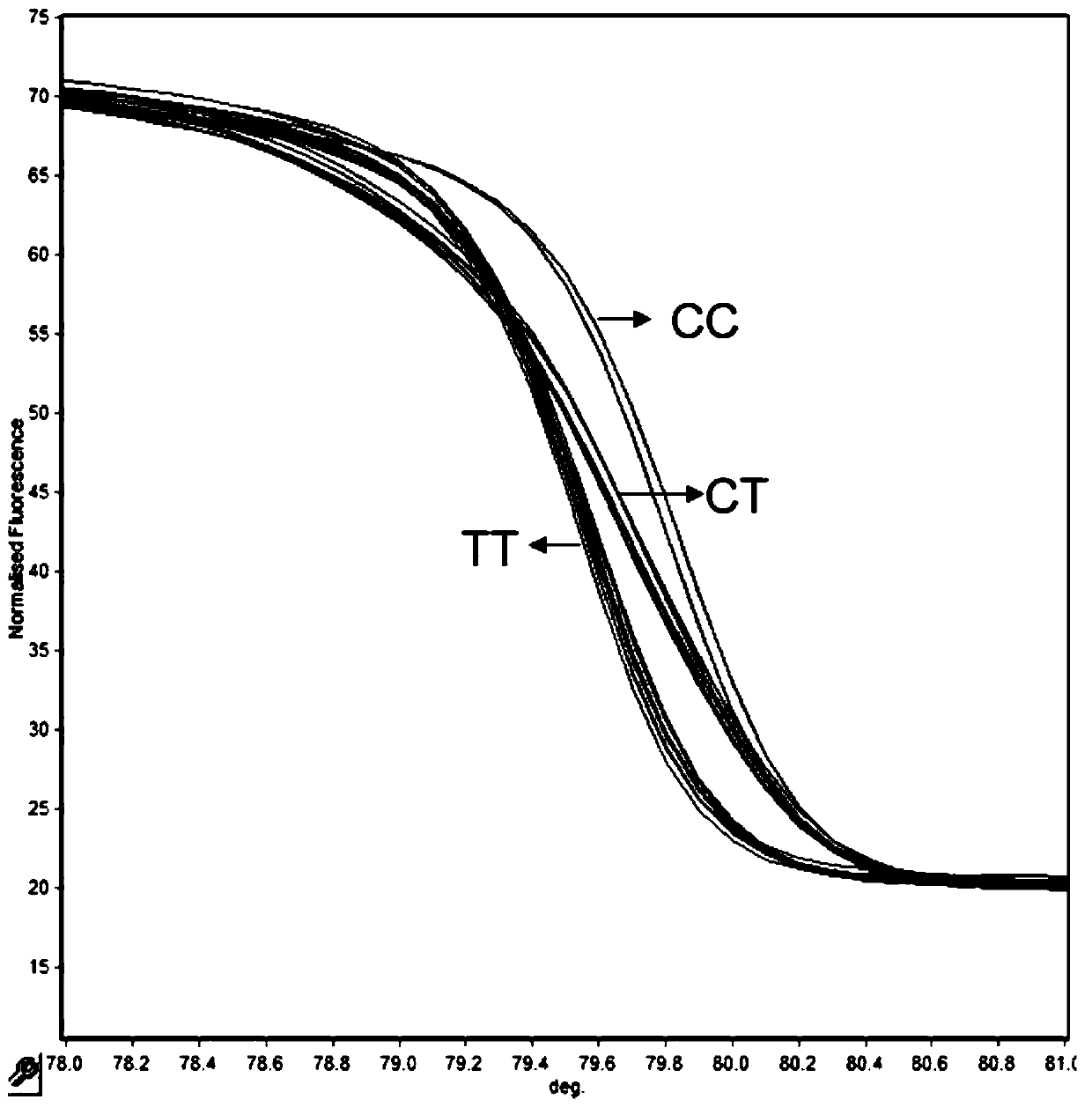
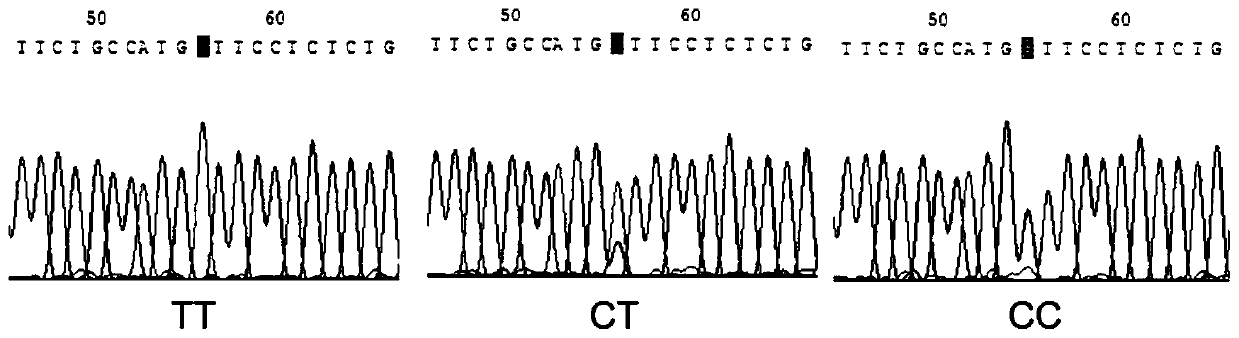
[0061] Example 3: Verification of the detection effect of the HRM typing primers used to detect the rs909253 site
[0062] The conventional rs909253 site amplification primers were used for amplification, and the length of the amplified fragment by direct sequencing of PCR products was 149 bp. The primers were synthesized by Beijing Dingguo Biotechnology Engineering Co., Ltd. The total PCR reaction system is 30 μL, and the reaction solution is: PCR Master Mix 15 μL, template DNA 1 μL, upstream and downstream primers 1 μL (10 μM / μL), add ddH 2 0 to 30 μ L, amplified on the PTC-100 PCR amplification instrument. Reaction program: pre-denaturation at 95°C for 5 min, cycle, denaturation at 95°C for 30 s, annealing at 62°C for 45 s, extension at 72°C for 30 s, a total of 35 cycles; extension at 72°C for 5 min, storage at 4°C. The amplified product was sequenced by Beijing Dingguo Biotechnology Engineering Co., Ltd. for the target fragment (reverse sequencing), and the corresponding...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com