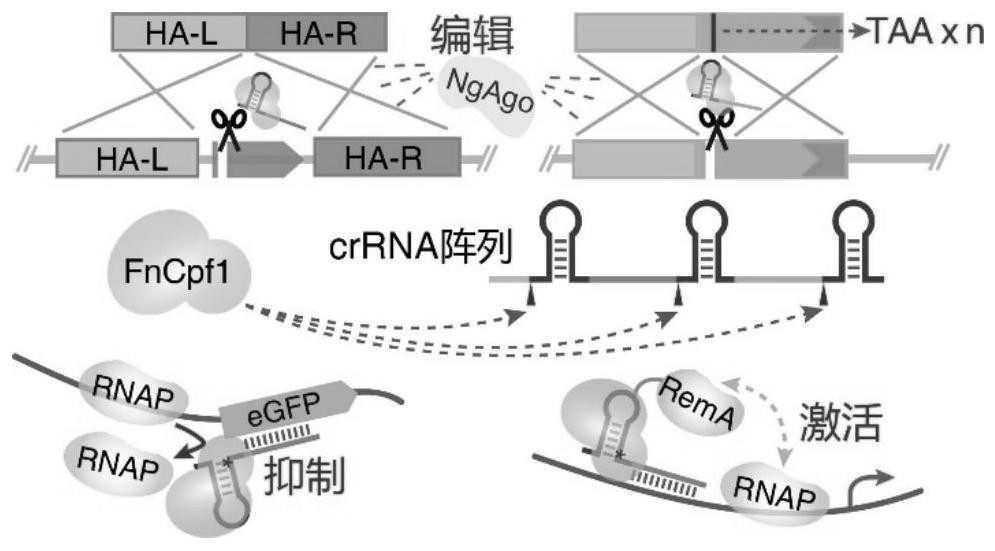
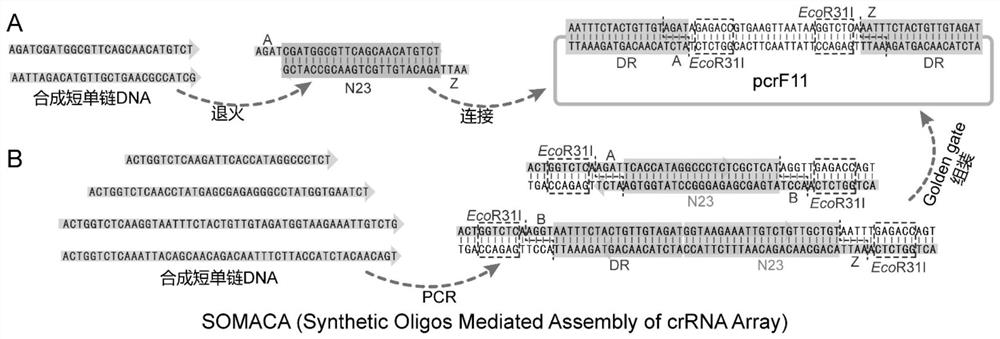
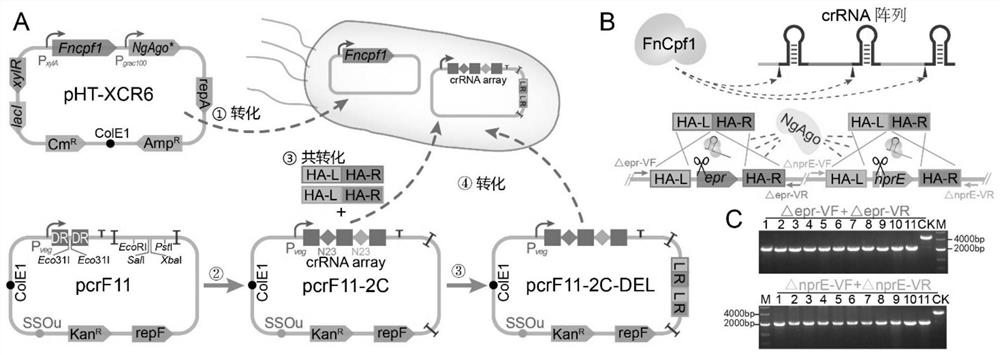
A Crispr Cpf1-based multiple gene editing and expression regulation system in Bacillus subtilis
A Bacillus subtilis, expression regulation technology, applied in the field of genetic engineering, can solve the problems of increasing the complexity of operations, reducing the stability of plasmids, increasing the difficulty of plasmid construction, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Construction of Cpf1 expression vector pHT-XCR6
[0044] Firstly, the plasmid pHT01 was linearized by PCR using primers with sequences such as 5'-ctgcagaacgctcggttgccgccgggcgttttttcgtcattcctctagagtcgacgtcc-3' and 5'-cgctcccttttccgttag-3' and the template DNA was digested with endonuclease DpnI; the plasmid pLCx-dCas9 was used as a template to amplify Xylose promoter PxylA; using PY001 as a template (from literature: Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, MakarovaKS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A, Koonin E V. Amplification FnCpf1 gene; finally, PxylA and FnCpf1 can be connected to the vector pHT01 to obtain the vector pHT-XC through the seamless cloning kit.
[0045] Then use primers with sequences such as 5'-ctaatgtcggattcctctaatcctctagagtcgacgtcc-3' and 5'-atgacgaaaaaacgcccg-3' to linearize the vector pHT-XC by PCR and digest the template DNA with endonuclease DpnI; amplify with plasmid pHT00 as a template IPTG-...
Embodiment 2
[0046] Example 2: Construction of crRNA array expression vector pcrF11
[0047] First, the synthetic DNA was used as a template to amplify the DNA fragment composed of the crRNA insertion region and the Bacillus subtilis replicon repF; then the kanamycin resistance gene was amplified by using the plasmid pUC57-Kan as a template; The DNA sequence was used as a template to amplify the fragment composed of the Bacillus subtilis single-stranded replicon ssoU and the promoter Pbs, and finally the plasmid pP43-egfp was used as a template to amplify the E. coli replicon ori; finally, the above-mentioned The four fragments were spliced to obtain the vector pcrF11 (sequence shown in SEQ ID NO: 2).
Embodiment 3
[0048] Example 3: Multiple gene editing in Bacillus subtilis based on CRISPR / Cpf1 system
[0049] During gene editing, pHT-XCR6 was first transformed into Bacillus subtilis, and the required crRNA array and homology arms were designed according to the needs of genome editing and added to the plasmid pcrF11 respectively. Here, the main six extracellular protease genes aprE, epr, nprE, bpr, mpr and nprB in Bacillus subtilis were selected as target genes for verification. It has been verified that if the homology arm is integrated into pcrF11, the complete deletion of two genes, the partial base mutation of six sites, or the insertion of a gene can be achieved at one time; and if the homology arm fragment is not inserted into pcrF11 , but when co-transforming with pcrF11 inserted with crRNA, it may be possible that due to the reduced transformation efficiency, only one gene can only be completely deleted at one time, or two sites can be partially mutated, and one gene can also be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com