Materials and methods for engineering cells and uses thereof in immuno-oncology
An engineering, cellular technology, applied in other methods of inserting foreign genetic materials, animal cells, anti-tumor drugs, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0832] The present invention will be more fully understood by reference to the following examples, which provide illustrative, non-limiting aspects of the invention.
[0833] The examples describe the use of the CRISPR system as an illustrative genome editing technology to create defined therapeutic genomic deletions, insertions, or substitutions (referred to herein as "genomic modifications") in or near target genes, which result in mutations in genomic loci permanent correction, or restore expression of target protein activity at heterologous loci. As described and shown herein, the introduction of defined therapeutic modifications represents novel therapeutic strategies that have the potential to ameliorate various medical conditions.
example 1
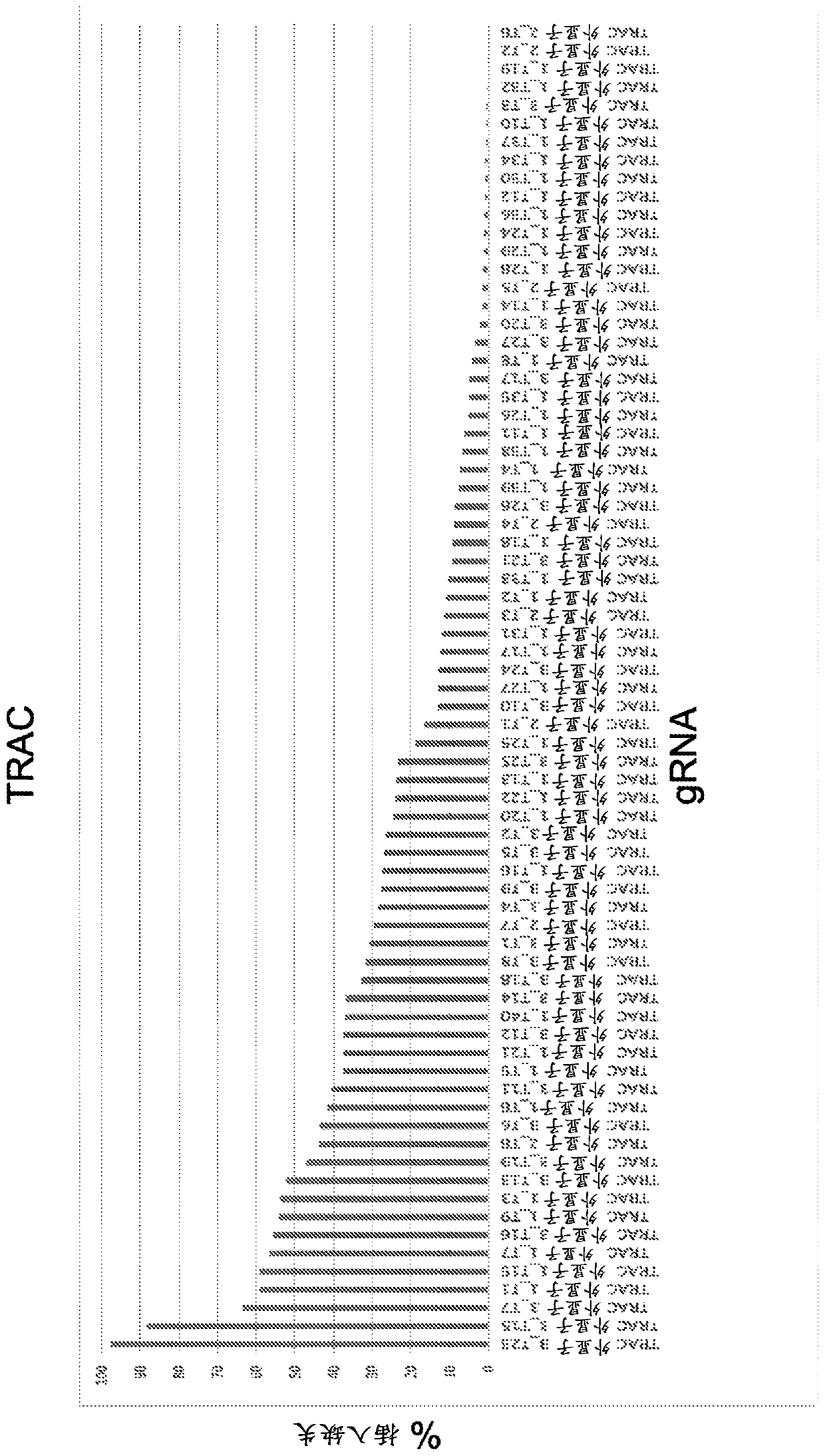
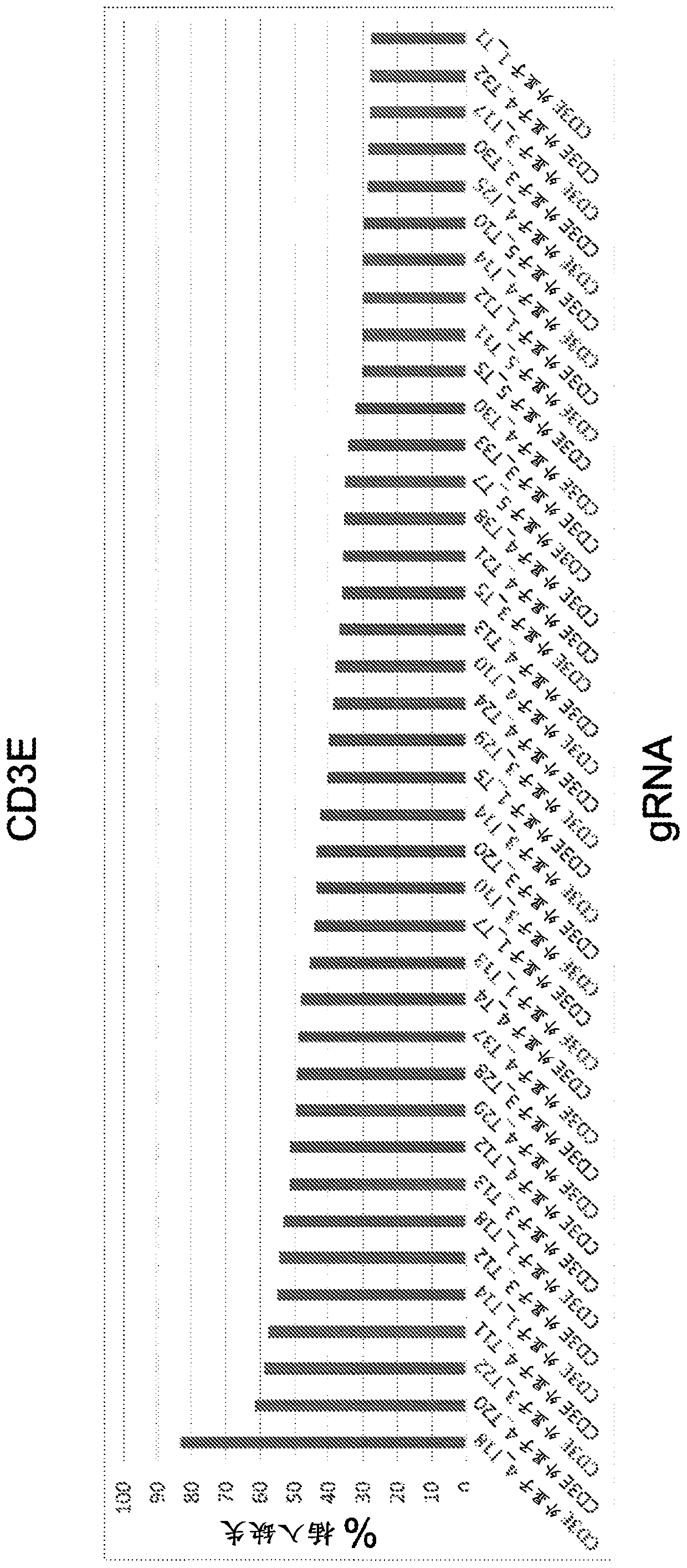
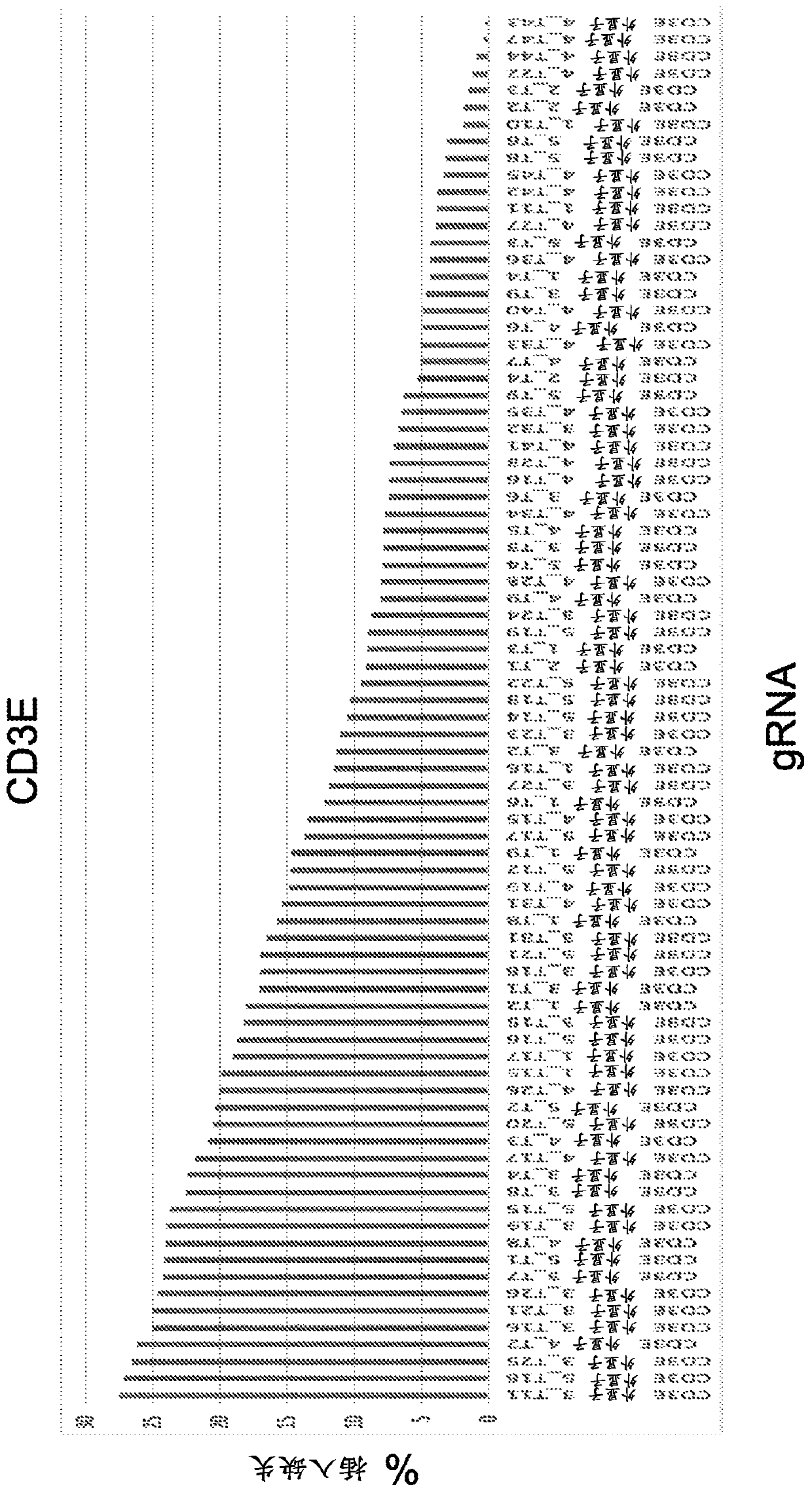
[0834] Example 1 - Screening gRNA
[0835] To identify a wide range of gRNAs capable of editing cognate DNA target regions, an in vitro transcribed (IVT) gRNA screen was performed. A spacer sequence is incorporated into the backbone sequence to generate a full-length sgRNA. Examples of backbone sequences are shown in Table 1. To generate a series of spacer sequences for gene disruption, each target gene (especially those containing the initiation ATG initiation codon and / or encoding key protein domains (e.g., DNA binding domain, Target genes for extracellular domains, etc.) select protein-coding exons. Submit relevant genomic sequences for analysis using gRNA design software. The resulting list of gRNAs was narrowed down to a list of ~200 gRNAs based on sequence uniqueness (screening only gRNAs that did not have a perfect match elsewhere in the genome) and minimal expected off-target effects. This set of gRNAs was transcribed in vitro and transfected into HEK293T cells con...
example 2
[0899] Example 2 - Gene knockout at the genotype and phenotype level in cells
[0900] This example demonstrates efficient knockdown by CRISPR / Cas9 of graft-versus-host (GVH) or host-versus-graft (HVG) genes or immune checkpoint genes at the genotype and phenotype level in primary human T cells remove.
[0901] Primary human T cells were isolated from peripheral blood (AllCells Inc., Alameda, CA) using the EasySep Direct Human T Cell Isolation Kit (Stemcell Technologies, Vancouver, Canada). cells at 0.5 x 10 6 cells / mL were plated in large flasks. Human T-activator CD3 / CD28 Dynabeads (Thermo Fisher Scientific, Waltham, MA) were resuspended and washed with PBS before adding to the cells. Cells were incubated with human T-activator CD3 / CD28 Dynabeads (Thermo Fisher Scientific, Waltham, MA) at a 1:1 bead-to-cell ratio in X-vivo15 hematopoietic serum-free medium (Thermo Fisher Shier Technology Company, Waltham, Massachusetts) supplemented with 5% human serum (Sigma-Aldrich (Si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com