Design method of nucleic acid capture probe for HLA typing
A nucleic acid capture and HLA-A technology, applied in the design of probes, nucleic acid capture probes for HLA typing and their design fields, can solve the problems of optimization, large differences, and failure to take into account, and achieve the effect of efficient capture
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Design of nucleic acid capture probes for HLA typing
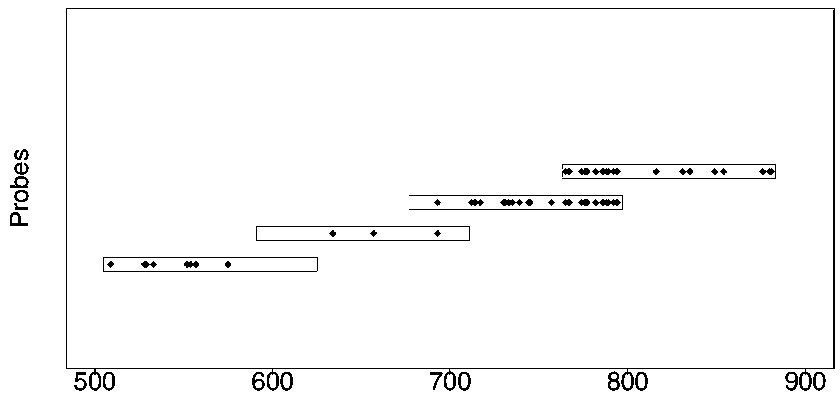
[0031] 1) According to the full lengths of the known subtypes of HLA genes (the length of most HLA genes is 3503bp), construct the sequence libraries of HLA-A, HLA-B, and HLA-C respectively.
[0032] 2) Use DECIPHER (from the R package of bioconductor) to perform multiple sequence alignments to find the main SNP sites on Exon2 and Exon3 of HLA-A, HLA-B, and HLA-C genes and within 100 bp upstream and downstream.
[0033] Taking Exon2 of the HLA-A gene as an example, the specific comparison steps are as follows:
[0034] ① Load the DECIPHER package——library(DECIPHER);
[0035] ②Use readDNAStringSet to read the fasta file of HLA-A——hla_A.fa;
[0036] ③Use the AlignSeqs function to align the read sequences;
[0037] ④Use writeXStringSet to write the aligned DNA sequence into a new fasta file, and the new fasta file is the result of MSA.
[0038] After alignment, a Multiple Sequence Alignment (MSA) alignmen...
Embodiment 2
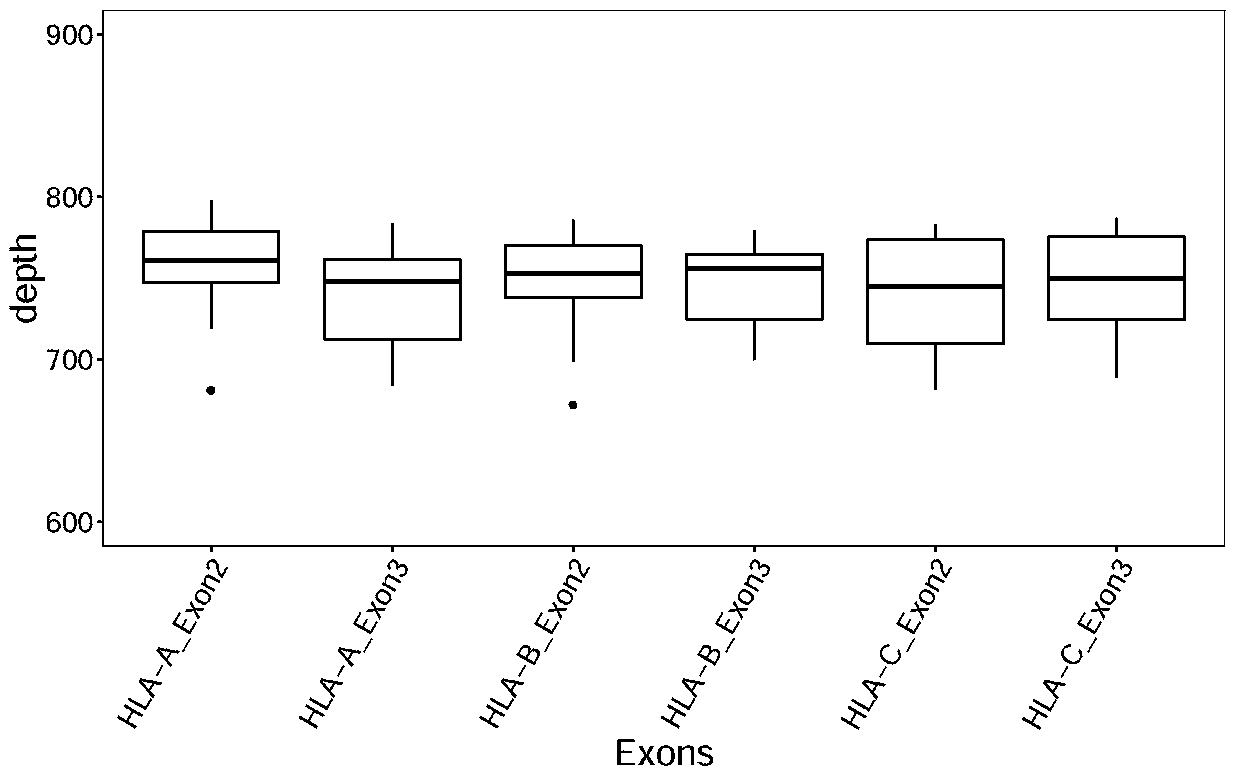
[0069] Example 2 Nucleic acid capture effect verification
[0070] DNA was extracted from 23 normal tissue samples, and the probes designed in Example 1 above were used for hybridization and capture of HLA genes. The library construction kits Accel-NGS ® 2S Hyb DNA Library Kit (Cat. No. 23024 / 23096) and 2S Set A / B Indexing Kit (Cat. No. 26148 / 26248) from Swift Company were used for library construction.
[0071] The operation steps of hybrid capture are as follows:
[0072] 1. gDNA fragmentation and purification
[0073] 1) Take 500ng gDNA according to the concentration of Qubit, add water to make up to 100μl, add covaris 130μl interrupted tube, set the program: 50W, 20%, 200 cycles, 330s. Take 1 μl after the interruption, and use the Qsep100 automatic nucleic acid and protein analysis system to detect the fragment distribution, and the main peak is 150-200 bp.
[0074] 2) Transfer the interrupted product to a new 1.5ml centrifuge tube, add 1.4 times the volume of AMPure b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com