Primer pair set for East Asian passiflora virus whole-genome determination and application of primer pair set
A whole genome and primer pair technology, applied in the field of molecular biology detection and identification, can solve the problem of inability to accurately identify viruses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
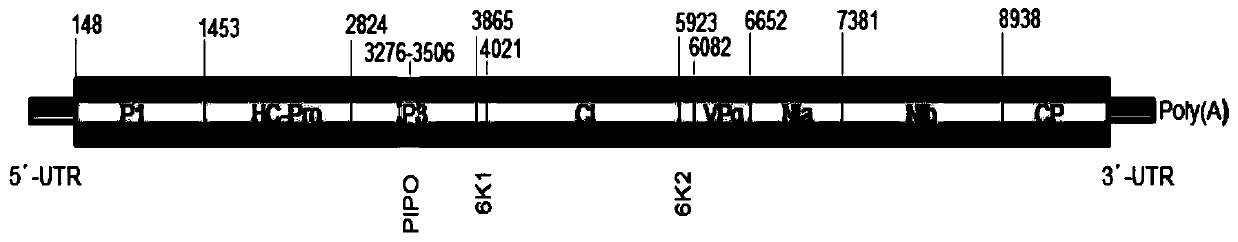
[0127] Embodiment 1, be used for the optimization of the design and the amplification condition of the complete set of primers that are used to amplify the whole genome of Passionflower virus in East Asia
[0128] One, be used for the design of the complete set of primer pair that is used for amplifying the full genome of East Asia passionflower virus
[0129] Utilizing the sequences amplified by general primers of Potyvirus and the gene sequence of East Asian Passiflora virus published so far, primers for whole genome amplification and sequencing of East Asian Passiflora virus were designed. The primer sequences are shown in Table 1.
[0130] Table 1. The whole genome amplification and sequencing primers of East Asian passionflower virus
[0131]
[0132]
[0133]
[0134] Note: F means upstream primer, R means downstream primer
[0135] After repeated design, testing and optimization, 7 pairs of primer pairs for cloning the whole genome of Passionflower virus in Ea...
Embodiment 2
[0149] Embodiment 2, East Asia Passiflora virus China Fujian isolate whole genome sequencing method
[0150] 1. Extraction of total RNA from samples
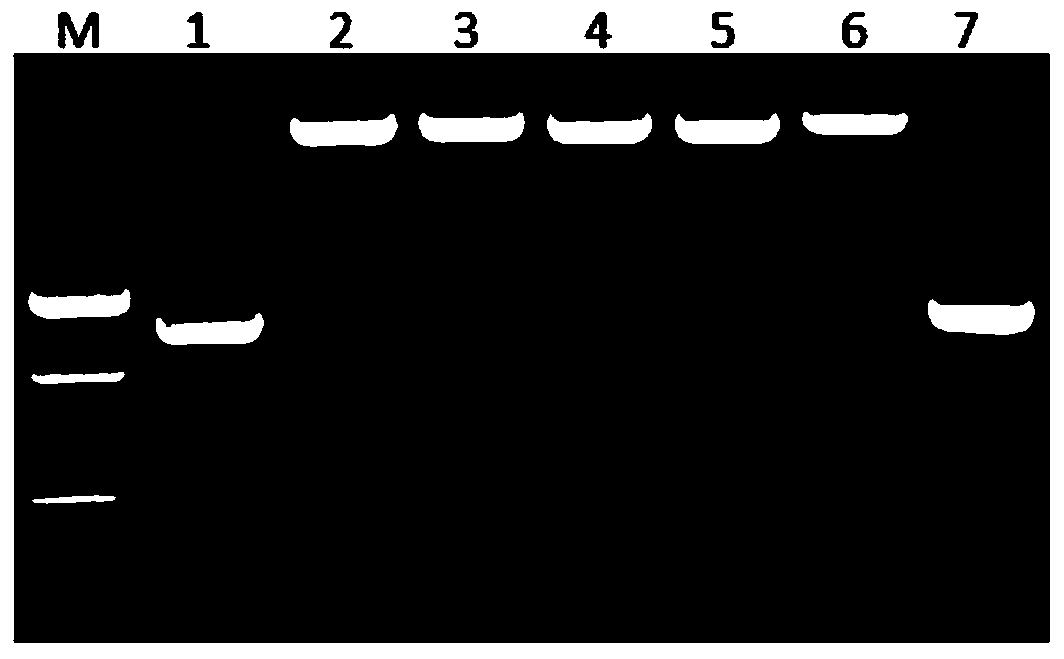
[0151] Take 0.1 g of passion flower leaves (Fujian, China) infected with East Asian passion flower virus, add TrizoL reagent, extract total RNA according to conventional methods, and detect by agarose gel electrophoresis, and obtain 50 μL of good quality total RNA.
[0152] 2. Whole genome RT-PCR amplification
[0153] 1) Reverse transcription: Take 3 μL of extracted total RNA as a template, add 1 μL of Random Primer, ddH 2 After 7 μL of O, first keep warm at 70°C for 10 minutes, then quickly ice-bath for 5 minutes, then add 5×RT Buffer 5 μL, dNTPs (10 mmol L -1 )2μL, RNasin (40U·μl -1 ) 1 μL and M-MuLV reverse transcriptase (200U·μl -1 ) 1 μL, and finally, cDNA was synthesized by incubating at 42°C for 1h and at 70°C for 10min.
[0154] 2) PCR amplification: with the cDNA obtained by step 1) reverse transcription as a templa...
Embodiment 3
[0164] Embodiment 3, the detection and the identification of Chinese Taiwan Passiflora sample East Asia Passiflora virus
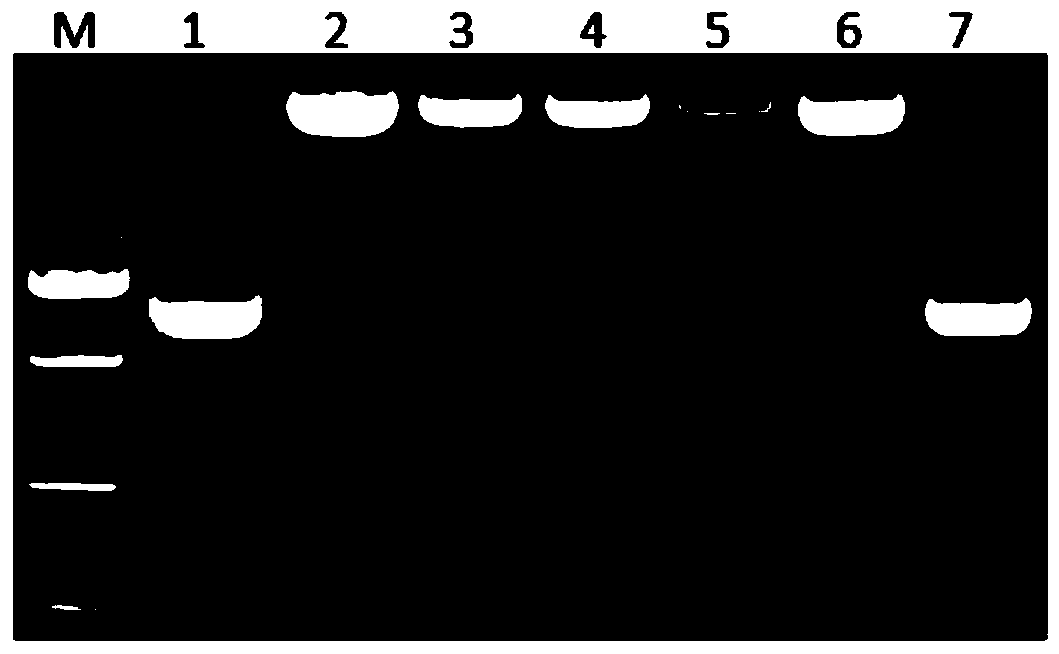
[0165] 1. Extraction of total RNA from samples
[0166] Take 2 samples (numbers TW-1 and TW-2) suspected of being infected with East Asian passionflower virus from Taiwan, China, as materials, weigh 0.1g respectively, add TrizoL reagent, and extract total RNA according to conventional methods. And detected by agarose gel electrophoresis, each 50 μL of total RNA with good quality was obtained.
[0167] 2. Whole genome RT-PCR amplification
[0168] 1) Reverse transcription: Take 3 μL of extracted total RNA as a template, add 1 μL of Random Primer, ddH 2 After 7 μL of O, first keep warm at 70°C for 10 minutes, then quickly ice-bath for 5 minutes, then add 5×RT Buffer 5 μL, dNTPs (10 mmol L -1 )2μL, RNasin (40U·μl -1 ) 1 μL and M-MuLV reverse transcriptase (200U·μl -1 ) 1 μL, and finally, cDNA was synthesized by incubating at 42°C for 1h and at 70°C for 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com