Rapid analysis method based on OrthoMCL clustering result
A rapid analysis and genome analysis technology, applied in the field of comparative genomics and bioinformatics, to achieve the effect of high versatility, high added value, and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
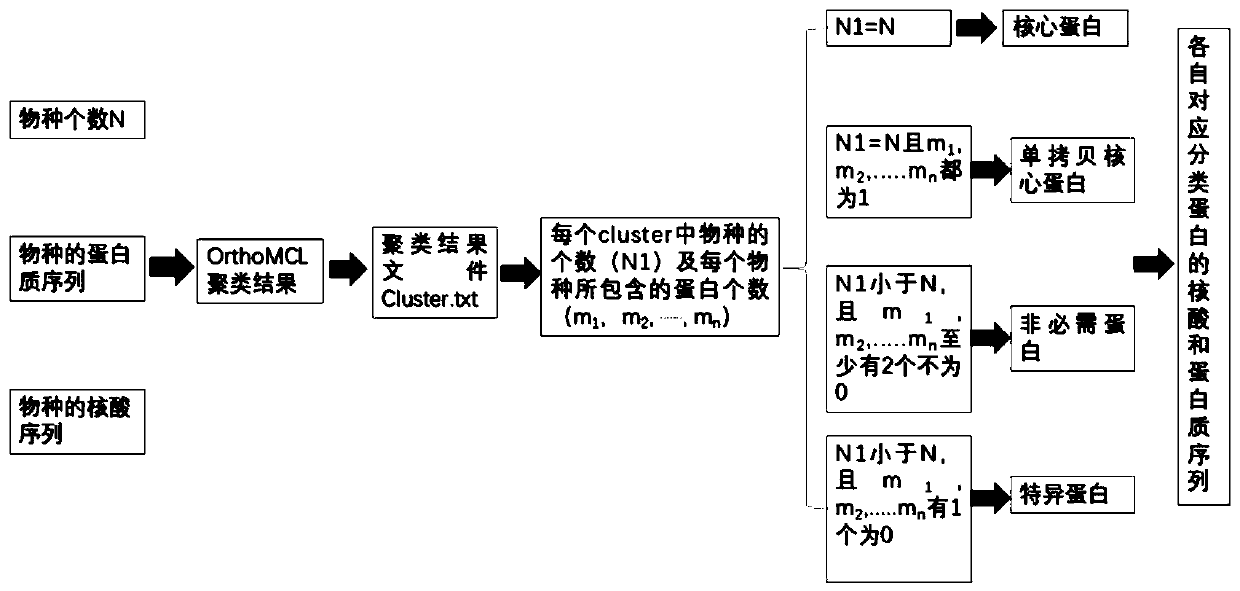
[0048] This embodiment provides a method for homologous clustering and rapid analysis of proteins based on the clustering results of OrthoMCL to analyze the proteins of 9 Trametes species, including the following steps:
[0049] Step S1, download the sequencing data files (including nucleic acid and protein sequences) of 9 Trametes species from the website of the National Center for Biotechnology Information (NCBI), and use the OrthoMCL clustering software to perform protein sequence analysis on the protein sequences of the 9 Trametes species. Homologous clustering, OrthoMCL clustering software can be downloaded from https: / / orthomcl.org / orthomcl / , OrthoMCL’s homologous clustering operation of proteins within a species is based on the existing technology, and will not be repeated here, obtained OrthoMCL clustering results;
[0050] Step S2, set the number of species used in the pan-genome analysis to N=9, count the number of species N1 contained in the cluster of each correspo...
Embodiment 2
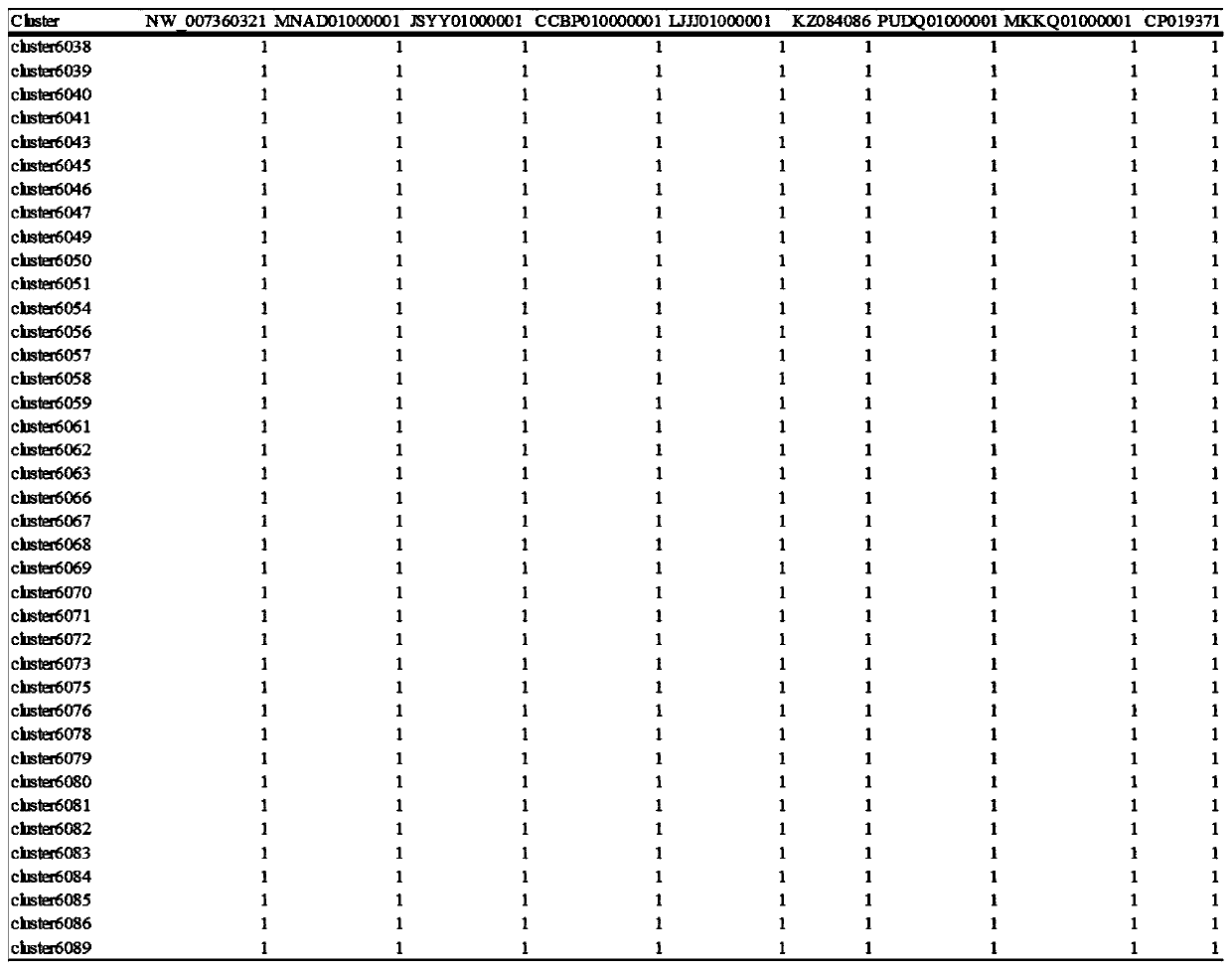
[0062] The only difference between this example and Example 1 is that the pan-genome analysis species are different. The files processed in this embodiment are the OrthoMCL protein clustering results of 9 strains of Veillonella atypica, the results are as follows Figure 4-5 as shown, Figure 4 It is the statistical result of the number of specific proteins in 9 species of atypical Veillonella, Figure 5 The homologous protein cluster60 in 9 species of atypical Veillonella corresponds to the protein sequence of each species. Based on the OrthoMCL protein clustering results of 9 atypical Veillonella species, after processing with this method, corresponding files can be provided for subsequent pan-genome analysis of atypical Veillonella species.
Embodiment 3
[0064] The only difference between this example and Example 1 is that the pan-genome analysis species are different. The files processed in this embodiment are the OrthoMCL protein clustering results of 66 strains of Porphyromonas gingivalis (Porphyromonas gingivalis), the results are as follows Figure 6-7 as shown, Figure 6 is a partial statistical result of the core protein of Porphyromonas gingivalis, Figure 7 The homologous protein cluster459 in 66 species of Porphyromonas gingivalis corresponds to the protein sequence in each species. Based on the OrthoMCL protein clustering results of 66 Porphyromonas gingivalis, after processing using this method, corresponding files can be provided for subsequent pan-genome analysis of Porphyromonas gingivalis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com