Primer group for detecting streptococcus salivarius, detection system and applications of primer group and detection system
A technology of streptococcus salivarius and primer set, which is applied in the biological field to achieve the effects of simple operation, easy promotion and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1 detection sample and bacterial classification
[0038] The strains required for the research are derived from the stool samples of the subjects. The subjects did not take antibiotics or similar drugs within three months before the collection of the stool samples. Refer to the instructions of the Changanyi ® stool collection box to collect the stool samples of the subjects. Store at 2-8°C in the laboratory.
Embodiment 2
[0039] Example 2 Primer Design and Identification
[0040] The microbial 16S rRNA sequence contains 9 variable regions and 10 highly conserved regions. In order to ensure the specificity of the primers, primers were designed in the conserved regions; in addition, compared with other variable regions, the V3-V4 regions can cover more than 98% of the Bacteria, and the length of the amplified product in the V3-V4 region meets the length requirements of qPCR for the expected product of the primer; based on this condition, a pair of primers that meet the requirements are designed to identify the total amount of all microorganisms in the stool sample.
[0041] Download the bacterial 16S rRNA sequence from the NCBI website (https: / / www.ncbi.nlm.nih.gov / nμccore / ) according to the bacterial name, and then use the primer design software Primer Primer 5 to design bacterial-specific primers; qPCR primer design requirements : The primer length is about 18-25bp, avoiding secondary structure...
Embodiment 3
[0045] The assembly of embodiment 3 kits
[0046] Streptococcus salivarius PCR reaction solution: Tris-HCl: 10-75mmol / L, pH8.5; Ammonium sulfate: 10-40mmol / L; MgCl 2 : 1-5.0mmol / L; dNTPs: 100-500μmol / L; final primer concentration: 0.25-1.25ng / μL.
[0047] Total bacteria PCR reaction solution: Tris-HCl: 50mmol / L, pH8.5; Ammonium sulfate: 25mmol / L; MgCl 2 : 2.5mmol / L; dNTPs: 350μmol / L;
[0048] Negative control PCR reaction solution: Tris-HCl: 40mmol / L, pH8.5; Ammonium sulfate: 30mmol / L; MgCl2: 3mmol / L; dNTPs: 300μmol / L;
[0049] Positive control PCR reaction solution: Tris-HCl: 40mmol / L, pH8.5; Ammonium sulfate: 30mmol / L; MgCl 2 : 3mmol / L; dNTPs: 300μmol / L;
[0050] Negative quality control: sterile water;
[0051] Positive quality control: Streptococcus salivarius genomic DNA.
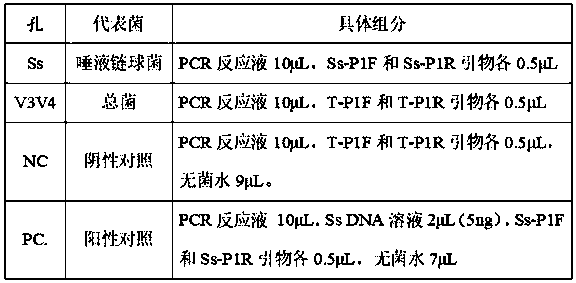
[0052] The structure of the intestinal microecology kit is shown in Table 2 below:
[0053] Table 1 Kit structure
[0054]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com